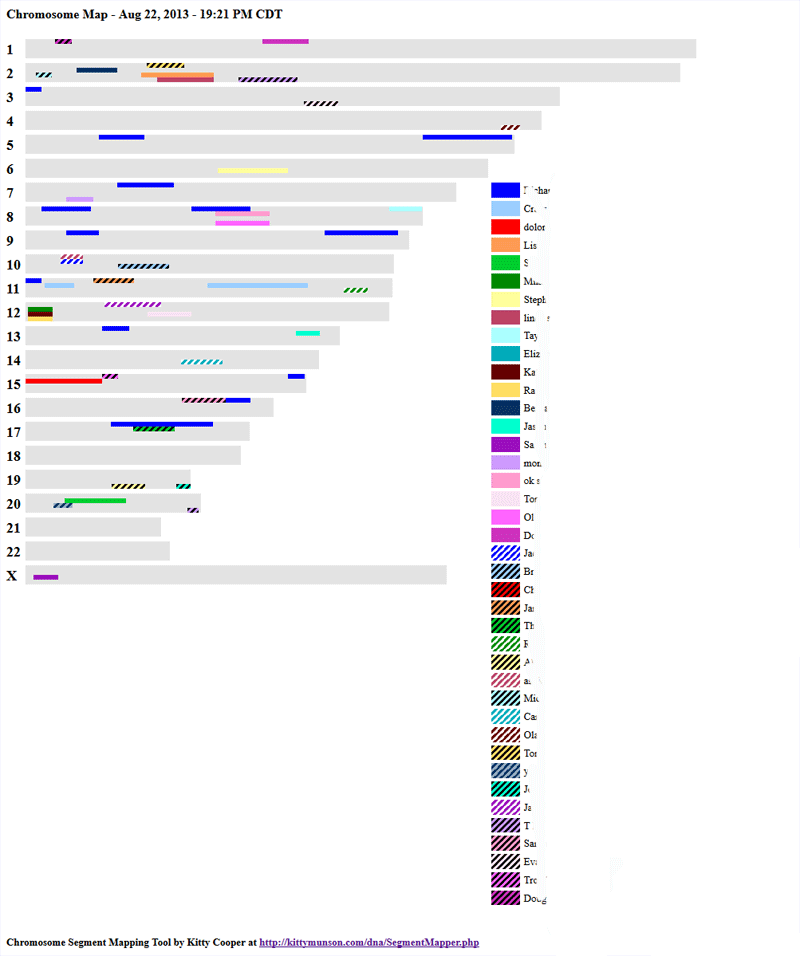
My tools for showing segments on a chromosome picture now allow you to pick the colors yourself. Here is an example of the ancestor DNA mapper with some very different color choices from the ones the program would use.
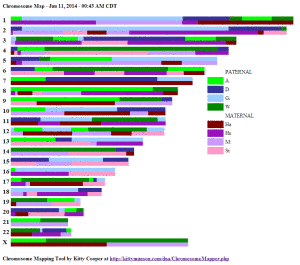 How to pick the colors is documented on the page for each tool.
How to pick the colors is documented on the page for each tool.
My ancestor mapping tool:
http://blog.kittycooper.com/tools/chromosome-mapper/
My segment mapper, up to 40 relatives
http://blog.kittycooper.com/tools/segment-mapper/
No changes were made to the one chromosome mapper so if you want color choice on that tool as well just let me know.
Another recent enhancement is that the segment mapping tool now allows you to specify as many lines as you like for relatives in your chromosome picture. See an example in my post about four generations of inheritance.