The current technology for personal DNA testing shows us the pair of values (A,T, G or C) from each of our parents at every tested position on a chromosome but cannot tell us what we got from which parent. If we could separate the DNA that we inherited from one parent, called phasing, and use that for DNA comparisons then perhaps there would be fewer false matches.
Phasing might bring matching segments smaller than 5cM into play. There have been many recent online posts and discussions among leading genetic genealogists about whether those small segment matches are real (IBD) or pseudo matches (IBC). Links to some of those articles are at the end of this article.
The X chromosome is particularly interesting for small segment comparisons because a male only has one of them to go with his Y, so we know all his X DNA is from his mother. Thus it is ‘phased’ to his mother’s results already. Perhaps then smaller matching X segments are more often real for men.
Dr. Kathy Johnston is a retired dermatologist who has been doing genealogical research for over 25 years and genetic genealogy for 10 years. She has been researching the X chromosome since 2008. She recently posted on facebook that small X segments can be IBD so I asked her for a guest post on the subject.
Read on to see what she has to say on this subject.
Males who have tested at Family Tree DNA have often asked, “Why do I have so few people on my X–match list but my mother (or daughter or wife) has so many?” Well, it could be that males are already phased which lowers the false positive rate significantly. Females have a lot of false positive matches on the X that include these small pseudo-segments. [Editor’s note: For a detailed discussion of false positives and IBD versus IBS see http://blog.kittycooper.com/2014/10/when-is-a-dna-segment-match-a-real-match-ibd-or-ibs-or-ibc/ ]
The X chromosome was added to the chromosome browser at FTDNA in January 2014 at the request of numerous customers. However, the X is not currently being used in the initial matching algorithm. One must have an autosomal match first at a significant threshold before an X match is called. Those who have any match on the X above 1 cM will be included in a list of X matching customers. It was apparent to me early on that this X match list for women was significantly inflated with false matches. It was never designed to be a true list, however, but rather a response to customer demand. The X match list for males is likely to be more accurate.
False negative matches are also a problem. That means you are missing a true match. You can have a significant match on the X which might not show up on your X match list at FTDNA because there was no autosomal match that met their threshold. AncestryDNA does not appear to utilize the X matches at all in their matching algorithm. However raw data from all three companies includes the X. 23andMe is the only company that uses the X chromosome in meeting the initial match threshold. An X-only match (without the autosomal match) is not very reliable in estimating a relationship but can still be quite significant [Ed note: see http://blog.kittycooper.com/2014/01/what-does-shared-x-dna-really-mean/ ]. Many X matches, which would not have been found otherwise, have been discovered because people uploaded their raw data from all three companies to GEDmatch.com.
It looked to me like the natural phasing of males created a significant gender difference in the X match lists at FTDNA. To test this hypothesis I looked at everyone who had ordered Family Finder (FF) tests through April, 2014 in our Southern California Family Tree DNA Project. There were more males (141) than females (110) who ordered the FF autosomal DNA test in our project. Males have traditionally tested their Y-DNA through this project because males were given group discounts; this may explain the higher number of males who ordered the autosomal test in addition. In general, there does not appear to be a significant difference in the company-wide autosomal testing of males and females but I could not obtain that actual number. When I looked at a comparison between the numbers of males and numbers of females in a majority of the autosomal match lists, there were no obvious differences. However, when I compared the specific X-chromosome-only match lists, there were striking differences between males and females so that is why I decided to painstakingly count these numbers in our project. Many more tests have been added in recent months but the following represents results from this initial study.
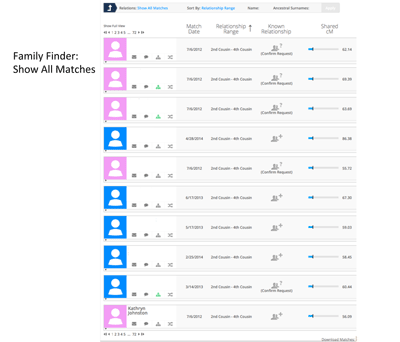 The graphics may have changed recently but a few months ago, the picture to the left was captured that shows a typical presentation of matches when looking at all autosomal matches for an individual. Females are in pink and males are in blue. Overall the percentage of females on a match list and the percentage of males were about the same for most customers. However since the default was blue, I had to account for several females who were mislabeled as males. I tried to balance out the testing company bias with the researcher bias in labeling gender. I made the best guess I could at the time by looking at names and tests that were ordered. I tried to document male or female whenever possible. I don’t think these biases really caused any significant change in the overall results.
The graphics may have changed recently but a few months ago, the picture to the left was captured that shows a typical presentation of matches when looking at all autosomal matches for an individual. Females are in pink and males are in blue. Overall the percentage of females on a match list and the percentage of males were about the same for most customers. However since the default was blue, I had to account for several females who were mislabeled as males. I tried to balance out the testing company bias with the researcher bias in labeling gender. I made the best guess I could at the time by looking at names and tests that were ordered. I tried to document male or female whenever possible. I don’t think these biases really caused any significant change in the overall results.
Females can expect to count at least twice as many total X matches because they have twice as many X chromosomes as males. However, the huge differences between males and females that I found in the list of X matches cannot be explained just based on the difference in numbers of sex chromosomes alone. Females had significantly higher numbers of people on their X match lists and the overwhelming number of matches were with other women.
The average number of Family Finder matches per “non-Jewish diaspora” male as of May 2014 was 436 in our project. Among these male project members, an average of only 1.3% of total FF matches also had X segments in common. Out of an average of 492 total matches for primarily non-Ashkenazi women, 21% of these were also considered X matches. Remember that the threshold was reduced to 1 cM for an X match so false positive IBS matches were to be expected.
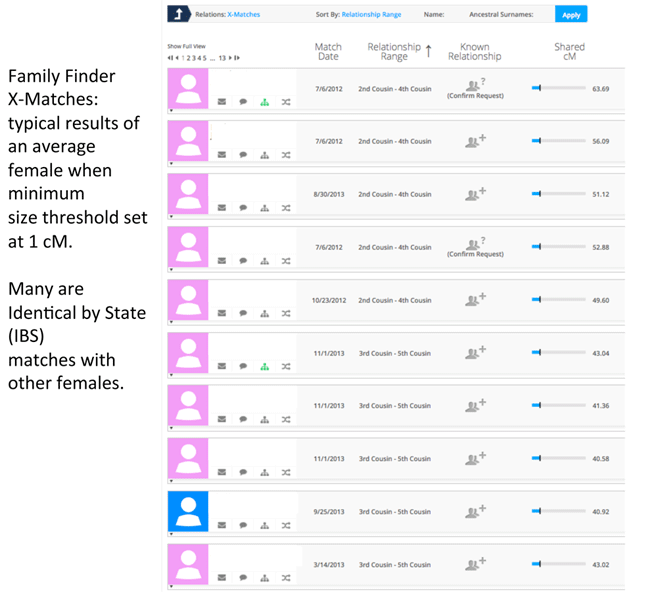
I estimated that the majority of genealogists in our project were representative of the American population of genealogists at large with a significant number having European heritage. Most people coming from minority groups had far fewer matches than those of European descent.
Only a handful of people had to be dropped from the final analysis because they were clearly the outliers. I subtracted out those with significant endogamous Jewish heritage who had > 1,300 total FF matches and over 30% Jewish Diaspora in myOrigins. This did not remove all people with Jewish ancestry, only those who appeared to me to fall at least 2 standard deviations above the mean. This was a common-sense approach and was not evaluated by a statistics expert.
Summary of the results for males:
- 6 % of men had no X-matches.
- 50% of men had only 1 to 5 X-matches. Many of these males only had X matches because they tested close relatives.
- However, some men had over 100 X chromosome matches and these turned out to be part of the Jewish diaspora.
- In the final analysis, in primarily non-Jewish males, an average of 1.3% of their autosomal matches also appeared on their X match list.
Summary of the results for females:
- The average non-Jewish diaspora female showed 105 (21%) X-matches out of 492 total autosomal matches.
- Of these X matches, the females outnumbered the males approximately 14 to 1.
- One female with over 1000 X matches was considered to be predominantly Ashkenazi in my estimation. She basically fell off the chart. I had no choice but to single out the Jewish population because the match lists were skewed as a result of very high numbers of matches within this particular endogamous population.
- Those females with over 30% Jewish Diaspora had female to male ratios of about 2.5 to one in their X match list, but were a very small group in our project.
In summary, males in our project on average had far fewer X matches than women, 1.3% versus 21% of total FF matches, unless they were from an endogamous Jewish population; in the latter case the gender differences were less significant but the matches were far more numerous. Most non-Jewish male genealogists had very few total X matches. I concluded that phasing of the data for males but not females had a large effect on these results.
When the threshold was lowered to 1 cM, it was no surprise that more than 1/5 of total matches were suddenly on a woman’s X match list. The X represents only one in 23 pairs of chromosomes, so with a higher threshold, there would not have been this high a ratio of X matches in women. What surprised me the most were the tremendous gender differences. The overwhelming majority of these matches among women were with other women.
I also wondered why were there so few X matches when a male was tested at a threshold of 1 cM? It must have been due to the natural phasing in men. In my estimation, only about 3% (2 times 1.3) of the total matches in women would have appeared on the X match list of women at or above 1 cM if females were also phased like males. Therefore I am estimating that the false matches outnumbered the true sequence matches by at least 6 to 1 in the females.
Just because a sequence can be called a true segment match does not mean that a match will have a findable common ancestor within the genealogical time frame. In this particular study, I was mostly interested in calculating the likelihood of a true X match based on the actual sequences in men confined to one chromosome rather than a true IBD match based on a pedigree analysis. This study made it possible to calculate the probable percentage of pseudo-segments in females caused by a lack of phasing assuming all other factors were equal. I would guess around 86% of the matches on a female’s match X list are not real when a threshold of 1 cM is set based on our project results.
In endogamous populations, there appear to be other factors besides a lack of phasing (such as low diversity, consanguinity, homozygosity, compounding of segments etc.) that create an unexpectedly high number of matches within the same population. FTDNA is known to have tested a large population of genealogists who self-identify as Jewish. It was my observation that MyOrigins could also identify them as being from the Jewish diaspora.
My intention was not to single out any particular ethnic group but rather to look only at the natural phasing in men that creates gender differences in X matching. The matching among endogamous populations needs to be studied further and could not be adequately evaluated in our project.
Would there be far fewer matches on the autosomes as well if everyone could be phased? Would phasing let us lower the threshold size well below 5 cM and still show significant true matching? I think so. This pilot study seems to suggest that the phasing of chromosomes is a way to weed out those false segment matches.
See the following blogs by Shannon Christmas, CeCe Moore and Blaine Bettinger for discussions of small matching segments in autosomal DNA:
http://throughthetreesblog.tumblr.com/post/105014866012/too-small-big-deal-size-matters-and-dna
http://www.yourgeneticgenealogist.com/2014/12/the-folly-of-using-small-segments-as.html/
http://www.thegeneticgenealogist.com/2014/12/08/small-matching-segments-examining-hypotheses/
Interesting. I can never include my stats into ANY type of analyzing because people always say that mine is unique. But I’m doing it anyway. Out of my 89 matches I get 49 who are X matches. My mother is 58% of X matches, not that much more than me. Not sure how to interpret that in this situation.
I found this very interesting and decided to check my husband’s matches. His father was Jewish, but his mother was not. Out of 28, there was only one other male. His match to the male was 4.55 cMs. His highest match was 4.97 cMs to a female with the same surname as the male. He had one 3.05 cM match. His other matches were smaller. Many of those were 2.16 cMs in the same place on the X. He also had several 2.1 cM matches in the same place.
Kalani your situation is unique but I enjoy hearing about it anyway. Endogamous populations have more X matches because of the intermarriage of their ancestors
My mother was half ashkenazi so my brother has 21 X matches at 23andme not including known family and all but 2 are larger than 8cM and at GEDmatch he has 212 X matches larger than 6cM (some are duplicated kits)
I would like your opinion on the relationship of an adopted “cousin” searching for biological parents. He has matched a person with 1458 cM and 32 segments. He’s pretty confident it is a half sibling. However, I have first cousins at 1039 cM and 37 segments and a half brother with 2023 cM and 42 segments. Another brother measures 1669 cM and 44 segments. Where do you believe this elusive person fits? Charts don’t seem to have a place for this match.
Autosomal DNA is not as precise as Y or mtDNA. The vagaries of inheritance are such that a few hundred CM plus or minus are within tolerance so half sibling fits.
The chart at http://www.isogg.org/wiki/Autosomal_DNA_statistics says that 25% or 1700.00 is about the expected amount for Grandfathers, grandmothers, aunts, uncles, half-siblings, double first cousins.
But send your DNA cousin to DNAadoption.com and to their yahoo group DNAadoption as well and ask there.
Pingback: Noli Irritare Leones » A round up post to remind you I’m still here
I have been hunting for a good article about low male x matches. Very helpful and interesting.
Thank you
Dear Kitty,
I am a 76-year old adoptee, born and living in SoCal, searching for my birth father. I connected with my Scot-Irish birth mother later in her life and she died never revealing the identity of my birth father due to painful memories. Around 6 years after her death I learned about autosomal DNA testing and had DNA testing with 3 different companies in 2013. From the results, I learned that my birth father was predominately Finnish, with a small amount of Volga-Ural Russian. Due to endogamy, I have around 2,000 Finnish DNA cousins, mostly 4th or greater, residing in Finland, with some third DNA cousins. Since my grandparents were most likely recent immigrants to the United States – probably 1890 -1910 – I have very few Finnish DNA cousins residing in the United States and those that I was successful im communicating with are unaware of my birth father.
I was hoping that I might identify my birth father in the 1940 US Federal census by entering on Ancestry all males born between 1918 – 1922, living in Los Angeles County, with parents born in Finland. Unfortunately, the 1940 census was the only recent census that didn’t inquire where the respondent’s birth parents were born, only the birthplace of the respondent. I went back to the 1930 US Federal census and there are almost 11,000 young men, nationally, born during that timeframe with parents born in Finland. Tracking them has been extremely challenging as many Americanized their first/surnames and many families just disappeared during those 10 years, perhaps due to relocations stemming from the Great Depression. I keep hoping for a Finnish 2nd cousin DNA match in the US with genealogical records to pursue. So far, nada.
My question is regarding the X chromosome: I have a female Finnish 3.9 cousin, residing in the UK, with total cMs of 62.2, longest cM 21.2, and X chromosome of 31.3 cM, longest cM of 15.8. Is this result meaningful and might I be related to this person through my birth father’s mother? If so, how might I proceed as there would be hundreds of descendants stemming from a common ancestor.
Thank you for any guidance you might suggest, as I’m not getting any younger. <3
Please go to DNAadoption.com and read their materials and perhaps sign up for a class.
Yes that woman is likely a 3rd cousin once removed or a 2nd cousin twice removed. Get her help and/or her tree and figure out which descendants of her gg-grandparents or g-grandparents were in L.A. at that time …
Nancy – one more thought, since there is such a large X match start by only looking at descendants who do not go through 2 males … in other words people on the X path of descent
Thank you, Ms. Kitty. I appreciate your feedback. Hugs…