Most of my matches at ancestry don’t see why they should upload their data to GEDmatch. I send them the URL of my slide presentation and extol the delights of the fun ancestry composition (admix) tools but it is hard to explain why I like to see where my DNA matches someone else’s. Curiosity? It’s fun? I love making these spreadsheets? Possibly it is because I am very interested in how DNA inheritance works and love to see which grandparent gave me which piece of DNA (n.b. it takes a lot of work to get to that point).
 When I know the common ancestor for a specific segment sometimes a new match fits in immediately to a family line. The best example of that is finding my previously unknown 3rd cousin Katy. When I saw where she overlapped I emailed her that it looked like she was related on my WOLD line to which she responded that her grandmother was a Wold. She has since sent me many wonderful family pictures that I had not seen before.
When I know the common ancestor for a specific segment sometimes a new match fits in immediately to a family line. The best example of that is finding my previously unknown 3rd cousin Katy. When I saw where she overlapped I emailed her that it looked like she was related on my WOLD line to which she responded that her grandmother was a Wold. She has since sent me many wonderful family pictures that I had not seen before.
Today I got an email from someone who had tested at ancestry and uploaded to GEDmatch. She wanted to know how to use my tools with her GEDmatch data. However my tools require a CSV file of overlapping segment data which cannot be downloaded in one fell swoop from GEDmatch, unlike at 23andme or Family Tree DNA.
Personally I built my many CSV files (one per person tested) slowly, as I compared each individual’s DNA results, contacted that match, and then cut and pasted the overlap information into my spreadsheets. Jim Bartlett did a great guest blog here on the process of building these DNA spreadsheets.
But I can understand the desire to see a quick picture of your matching DNA. GEDmatch does have a chromosome browser where you can see the overlaps, although the presentation is somewhat different from other sites. A little known secret is that you can massage that function’s table output into a spreadsheet (see end of this post for the technique).
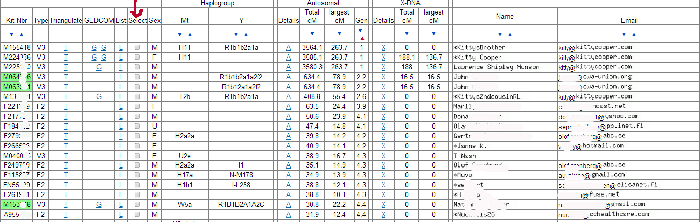
To use the GEDmatch chromosome browser you start by clicking on the one-to-many under “DNA analysis” on your GEDmatch home page (click here for that slide) then enter your kit number on the form on the next page. After a bit of a wait you will be presented with a large table that is a matrix of the information for your matches.
By the way, these column headings are explained in the handout for my slides, as well as in the GEDmatch utilities manual. Both of these are in my download area.
Now to get a chromosome browser view from the one-to-many results page, you have to click on the box in the “Select” column for each person you want to see in your chromosome picture. For example you might pick your top forty matches. Then every chromosome is shown separately on the resulting output page. Here is how chromosome 1 from my Dad’s top ten looks (click the image for a larger version):
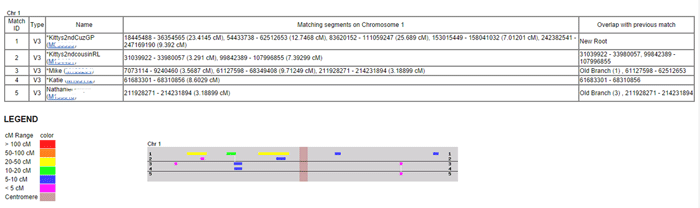
Each person is represented by a line in the table and chart. So in this example, line 1 is my second cousin RL. The colors used indicate the size of the match in centiMorgans (cMs – click here for the definition). This is useful because the display is in base pairs, which is a real physical distance, unlike the cMs, but is not as representative of the degree of the match as the cMs are. So anything colored green or warmer is a good match. I tend to only pay attention to the blues when they overlap other matches and I ignore the purples.
When the segments are overlapping with another person in the group this is shown in the far right column with the start and stop points of the match in base pairs. So GP and RL are overlapping segments on the chart as indicated in that column. This DOES NOT mean that they match each other there, although that is expected since they are 2nd cousins to each other as well (GP is actually a 1st cousin to me). A one-to-one comparison for these two would tell me that they match. Remember the reason this is called a 2D chromosome browser is that everyone is being compared to just one person, not to each other.
When a DNA segment does not overlap with the person in the line just above it, then the match up is shown with a white vertical line on the chart to the other person’s horizontal line and the notation Old Branch(#) is used in the far right column. The number (#) in parenthesis indicates which line number the matching person is on. So Mike on line 3 has a small overlap with RL on line 1 and his daughter Katie has that same matching segment on the next line down. The spot where Mike’s cousin Nathaniel on line 5 matches him is indicated by the words Old Branch(3) followed by the numbers indicating where the segment starts and ends. Mike, Nathaniel, and Katie are not related to RL only to GP, according to the known paper trail.
Why might overlapping segments not be a match? Because there are two sides for every location on these charts, since our DNA consists of 23 paired chromosomes, each of which is a recombined mix from one parent (except the Y). For more on that concept read my write-up of DNA basics on this blog; it has a number of links to other good pages with explanations of the basics as well.
Click here for an even more detailed write-up of using the GEDmatch Chromosome Browser for triangulation by Sue Griffith.
Now back to the original question. David from DNA-NEWBIE described massaging the chromosome browser output in a post in that mailing list, but then referred me to a wonderful document from the DNAadoption.com folk that shows how to do it step-by-step:
http://dnaadoption.com/uploads/DNAadoption/DNAadoption_files/DNAadoption/Combining_Results.pdf
Also there is a terrific all-in-one tool called Genome Mate that will give you overlap pictures of your overlaps from your ancestry data that has been uploaded to GEDmatch. I have not yet used it myself but I hear good things about it.
So dear cousins from ancestry, does this help? Let me know how I might improve this write up.
UPDATE: 20 Mar 2017 – I prefer to use the Matching Segment Search to create the first spreadsheet for a cousin tested at ancestry. See my post called “Taking it to the next level – DNA spreadsheets.” Also when I first wrote this post, I thought that the overlaps shown were actual matches to each other but they are not necessarily. They are just where those people match the original person being compared.
Of course we in the DNAAdoption community are already working on the tools to automate the manual steps of Combining the results. If we could push Gedmatch to provide CSV it would go even faster.
Just dipping my toes in the whole DNA comparison stuff. The PDF that you linked to is GREAT – I am on the cusp of understanding all of this, but am wary about making incorrect conclusions and wasting valuable time by not setting things up in a relatively (<- See what I did right there?) streamlined manner. It is a tall ask, but I'd would be great if someone did a screen cast of the basic steps in
1 – identifying a match in FTDNA/23andMe/Ancestry/GEDMatch/etc
2 – Exporting those match(es)
3 – importing to excel
4 – manipulating the data (NOT and excel tutorial, but rather manipulating the data and WHY you are doing that)
5 – understanding the results and then
6 – Triangulation
It's a lot, but I think someone with knowledge and a little technical prowess can record that in a 10 min segment (maybe 2 @ 10 min segments) that can be used for reference for everyone who follows.
Joe, I think this old post of mine covers most of what you ask for
http://blog.kittycooper.com/2013/01/finding-distant-relatives-with-autosomal-dna-testing/
and perhaps if you read Kelly Wheaton’s lessons that will help too. There is no one size fits all for DNA research, a lot depends on your specific goals.
You might also have a look at genomemate which looks to be a good tool for organizing this data with step by step help. I am experimenting with using it for my Aunt and will blog about it someday soon.
Hi Miss Kitty,
How can I upload my DNA files from Ancestry? Did you know Ancestry is shutting down some of the DNA results they have now? We have until the end of this month so I need to get it off. Is it costly to upload?
Janice
Almost forgot, a quick post or information about a hierarchy of what you should be focused on when looking for matches.
# of segments
Matching Segment Length
Total (SUM) of all Matching Segments
Let’s talk about this.
Case A: 1 matching segment, and that segment is 90cM
Case B: 2 matching segments, largest is 30cM, total is 90cM
Case C: 15 matching segments, largest is 11cM, total is 90cM
Is one better than the other? Why/why not.
Janice –
Only the Y and the mtDNA are being shut down. It is cheap to transfer the Y to ftDNA which has wonderful tools and many surname and locality projects for Y DNA – here is my affiliate link to buy that transfer: Click here to transfer your Y-DNA results to Family Tree DNA
There are also many good blog posts on what to do about this – see what Roberta Estes says about this on her blog – http://dna-explained.com/2014/06/11/transfer-dna-results-or-retest-at-family-tree-dna/
The tag spreadsheets here will get you the Jim Bartlett article linked to from this post as well as my older simpler article http://blog.kittycooper.com/2012/11/making-a-spreadsheet-of-your-dna-matches/
These may answer your questions. Also there is lots of stuff at the DNAadoption site.
As to the hierarchy of what to focus on, large segments are good. More than one good sized segment is good. Case A and B are effectively the same since A is probably more than one segment that just happen to be adjacent. Both look like a 4th cousin. In case C, I would want to know how many segments > 7cM and whether it was from an endogamous population group.
Autosomal DNA is a bit amorphous and random so there are no easy answers
No easy answers? I want my money back!
Seriously, thanks for the guidance and I am reading (and re-reading) all the info that you have posted above. Almost. There.
Hi Kitty
I have tried gedmatch and have about 20 people who have over 10cm with me , 2 of these people are in a single block, 12.6cm and 11.8cm.
my question is – they sit on chr19 and chr20, do these Chr areas mean anything?
I was told Ch1 is based on your grandparents and chr22 is your very ancient details. I do not know of what the other Chr mean.
kind regards
vic
Vic –
Nice to have 20 matches of over 10cM! When you have only one segment > 7cM the relationship can be as recent as a 4th cousin or as far 14th cousin … see http://blog.kittycooper.com/2013/02/largeibdsegmen/
The 23 pairs of chromosomes are far more complicated than the simplistic view you were given by whomever. My post on a 4 generation inheritance chart done by Angie Bush shows which segment her daughter got from which great grandparent http://blog.kittycooper.com/2014/09/using-the-chromosome-mapper-to-make-a-four-generation-inheritance-picture/
Wikipedia has good descriptions of each chromosome, here are the ones your mentioned:
http://en.wikipedia.org/wiki/Chromosome_20_%28human%29
chromosome 20 has been updated to just over 63 million base pairs and contains 897 or 1,068 genes
http://en.wikipedia.org/wiki/Chromosome_19_%28human%29
Chromosome 19 spans more than 59 million base pairs and contains contains 2,072 or 2,670 genes
http://en.wikipedia.org/wiki/Chromosome_1_(human)
Chromosome 1 spans about 249 million nucleotide base pairs and is currently thought to have 4,316 genes
Thanks Kitty for great links.
I think its strange that I get .20% in 23andme with a person who is related to me via BDM’s and is my great-great grandmother ( on my maternal side). She was born in 1811.
Yet others who are higher in % I have not found via BDM from present to year 1700 ( must be a minor branch of the main branch )
I will try your Chr-mapper………..MyHeritage has its limits
all the best
vic
You are welcome.
What does BDM stand for Vic?
Once you get past 3rd cousins the amount of DNA inherited is more and more random. My Dad shares .20% with one 3rd 1R but only .09% with her brother and .11% with her sister. I find the number of large segments is a better indicator of a close relationship than percentages.
hi
BDM = Birth, Death and Marriages ( registar records, hand written, no search engine……pain in the ### to find relatives.)
Time and lots of effort, but at least I tracked a solid line to 1680 paternal.
regards
hi there, i am brand new to the DNA testing and just got my results back from ancestry and uploaded to gedmatch.com my question is this, i have a brick wall on my fathers side but have a few possible branches, i would like to compare my DNA results with others with the same surname. to do this i need their kit numbers of course, do you know of any website that lets people list their kit number and surname so others can use that to compare DNA?
Michael –
For surname and straight paternal testing, the best tool is a Y DNA test rather than an autosomal test. See http://www.isogg.org/wiki/Y_chromosome_DNA_tests
So if you can find a male line descendant of your brick wall get his Y tested at Family Tree DNA – also there are many projects for specific surnames that are Y tested over there.
That being said, try the message boards for the surname you are interested in at rootsweb and post this query there – http://boards.rootsweb.com/?o_iid=33216&o_lid=33216&o_sch=Web+Property
A few more thoughts Michael.
To use autosomal for your brick wall it can be very helpful to get 1st and 2nd cousins to test as they isolate which family lines a specific segment may come from. So test a few cousins that both descend and do not descend from your brick wall.
I solved the brick wall in our family using Y with the help of a few Norwegians, one of whom found a candidate to test whether our theory was good see
http://blog.kittycooper.com/2013/03/we-have-found-our-ancestor-lars-monsen/
and the follow up
http://blog.kittycooper.com/2013/05/its-a-match-lars-monsens-ancestors-are-found/
Hello, I have a mach on 23andme predicted as 3rd with 2nd to 4th range. Her and i share 1.27% on 5 segments. Now on gedmatch is shows gens to mrca as 3.5. What does this suggest?
If it helps, her daughter on 23andme shows as 3rd cousin (to 4th range) and we share 0.97% on 4 segments.
Raiza –
Autosomal DNA is not precise due to the randomness of DNA inheritance so no one can give you a firm estimate. The Gedmatch 3.5 generation estimate is 2nd-3rd cousin (parent/child=1,1st cousin=2, 2nd cousin=3).
Look at the total cM as well as percentages and compare them to this chart:
http://www.isogg.org/wiki/Autosomal_DNA_statistics
Also look at how many large segments. Plus if you are from a very intermarried population such as Polynesians, Ashkenazim, or Mennonites the estimates can be misleadingly closer that they really are.
You will want to look among your g-grandparents and gg-grandparents for your shared ancestral couple.
Hi. I am sorry to bother you but from the look of your blog you seem to be the person to ask!
I have just today uploaded my Ancestry DNA test results to Gedmatch and whilst waiting for the processing to finish (not daring to go elsewhere), I saw my 23 Chromosome data appear. Or rather, what I took to be the data. To say I was surprised by what I saw would put it mildly. Do you know if I was seeing things correctly? Did I see my sex chromosome data?
Thank you for your patience.
Anne
So are you saying you also tested at 23andme? I am not sure what you question is but I really recommend you join the yahoo mailing list DNA-NEWBIES or the facebook group Ancestry-Gedmatch-FTDNA-23&me-Genealogy and DNA and ask your question at one of those places. I am on vacation in Norway!
My DNA is not linking to my dad or grandmother (his mother) and when I uploaded to GEDmatch it states there is “no DNA match.” I also compared my son’s DNA. I was told that my mother’s DNA can suppress my father’s DNA. I’m totally confused. I’m starting to believe he is not my biological father. Please help!
Your DNA will always match each parent on every chromosome for the whole chromosome except a few hiccups where there may have been small errors. You will match a grandparent at about 25% but it can be smaller or larger.
Double check with the company you tested yourself and him at that there was no error and that you downloaded the correct raw DNA for yourself and your Dad.
If all is correct then sorry, he is not your biological father.
One proviso, male children will not match their father on the X since they got a Y instead
So glad to find someone who is trying to make sense of the GEDmatch technology. I’m a relative newcomer, so confusion is my name.
Right now, I have a giant GEDmatch – Matching Segment Search – V2.1.2 –
Using nfs kits spreadsheet and I can’t figure out how to know which color band means exactly what. Is it keyed to various part of the planet? And if it is, WHERE is this information. I wouldn’t mind if GEDmatch included table of contents and may … an index? Or glossary? Something?
I was really happy to find your post! I have a blog too, incidentally at
https://teepee12.com/ so if you think I’m weird, really, I’m not. I’m just a blogger trying to dope out how GEDmatch works in hopes of actually making sense of the DNA I got from MyHeritage where they explain nothing without a pre-payment.
Best,
Marilyn
GenomeMate.org appears gone, replaced by
https://www.getgmp.com/
Pingback: Tuesday’s Tip: GEDmatch.com – Copper Leaf Genealogy