When I first started doing DNA testing and I saw that full siblings shared only about 2800 cM of their DNA, I was surprised. I expected more. Then I thought about it. We have 23 pairs of chromosomes but the testing process cannot separate the two parts of each pair so our matches are seen as if we had only 23 unpaired chromosomes.
The matching segments are listed and totaled as if there was just one side of each pair even though it is using the data from both sides for the comparison; that is why they are called half identical regions or HIRs. Click here for my blog post with a deeper discussion of why doing comparisons that way can create false matching segments.
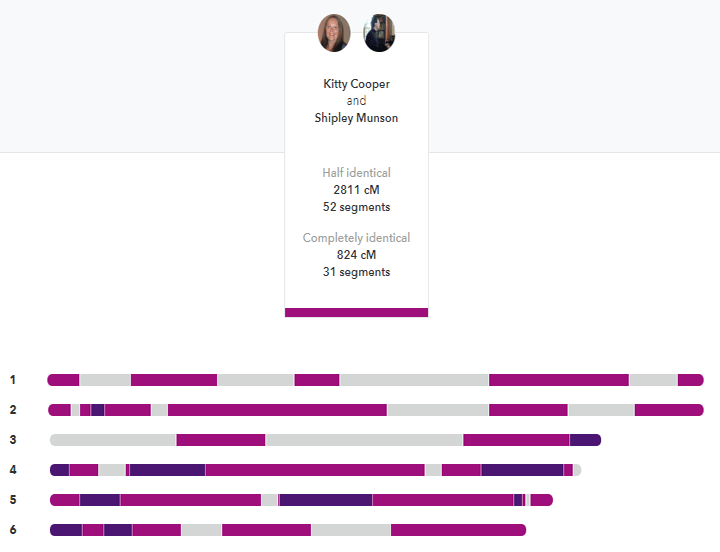
My brother and I compared at 23andme. The darker segments are the FIRs. Click it for the full image.
Siblings are usually listed as sharing about 50% of their DNA but that is only the half identical part that most companies measure. How many places did they get the same DNA from both parents on both pairs of a chromosome? In other words how many fully identical regions (FIRs) do siblings share? In my experience, they share about 800 cM or so that are fully identical. Thus the total of the FIRs and HIRs is about 3500-3700 cM; the same amount expected of a parent match. That intuitively makes much more sense to me.
So what would be the logical basis for this? Each parent gives each child half of their DNA but not all of it will be the same half. Roughly half of what each parent gives me will be the same as what my brother got so about 2 times 1800 for 3600; yet most full siblings are shown as sharing only about 2600-2800 cM of half identical segments. The rest of that 3600 is found in the fully identical segments.
A recent query I got was “I only share 2553 cMs with my sibling are we full or half?” By the online calculator at DNApainter they are full siblings sharing a low amount, but another possibility is the father of one sibling was the brother of the other. That is called ¾ sibings. To tell the difference total up the FIRs and the HIRs. If they total less than 3000 rather than the 3600 for full siblings then they are most likely three quarters siblings or some similar relationship.
23andme totals both the FIRs and the HIRs unlike any other testing company, as shown above, however those totals include the X which throws it off a bit for my calculations.
 Wherever you tested, you can get the FIRs and the HIRs by comparing the two kits at GEDmatch.com.
Wherever you tested, you can get the FIRs and the HIRs by comparing the two kits at GEDmatch.com.
Here’s how. Presuming you have both uploaded to GEDmatch (and hopefully both opted in to helping law enforcement identify violent criminals and victims – click here for my post on that), start with a One-to-One comparison of the two kits.
Continue reading →