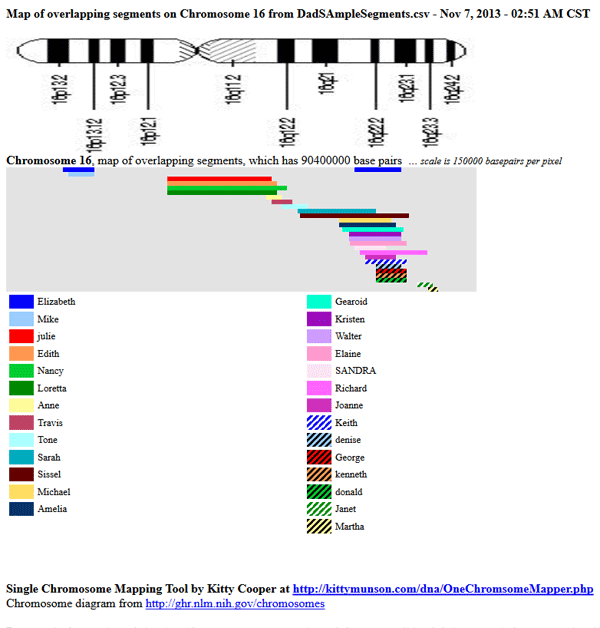
Sometimes you might like to see a detailed picture of all the largest overlapping DNA segments on one chromosome. If that is the case, then try my One Chromosome Mapper tool at (the secure url is no longer working): http://kittymunson.com/dna/OneChromosomeMapper.php.
This tool will show you up to 40 people’s overlapping DNA segments on the specified chromosome. As with my other tools, best to sort your CSV file by the largest segment. Then delete the lines with very small segments (less than 3cM) and then resort by start point to get the cascading effect in my sample above.
Note that there are popups that show the exact locations and cMs of each segment on the actual output. The NIH.gov chromosome diagram is to give a reference although the scale may not be perfect you can see when a match crosses that central bit called the centromere which means it may really be two segments.
As with my other tools, the column headings in your data file have to be exact (see below), although several variations are allowed, and the CSV format has to be PC compatible. If you are on a Mac please check the how to make a CSV article since this can be a problem. You can use the CSV file generated from countries of ancestry at 23andme or the FIA file from DNAgedcom.com without changes, although as always I recommend sorting by largest segments
The One Chromosome Mapper requires the column headings and contents as listed below, case sensitive, in any order, in your CSV spreadsheet
Current List of Required Columns in your CSV file for this Tool
| Column name | Alternate Name | Must Contain this Data |
|---|---|---|
| Comparison | MatchName or any column name you specify |
This is the name to use in the chromosome map picture, you can tell the program to use a different column name for this. |
| Chr | Chromosome | Chromosome number 1-22 or X |
| Start | Start point SegmentStartInMegaBasePairs |
The starting number for the segment location used by 23andme and Family Tree DNA, in the 23andme ancestry these numbers are divided by 100,000 so check the box for decimal if you are using that style of number. |
| End | End point SegmentEndInMegaBasePairs |
The ending number for the segment (see above comment). |
| cMs | Genetic DistancecentiMorgans (cMs)SegmentLengthInCentiMorgans | The number of centimorgans in the segment, used for display purposes only. Since this is for display only you can put anything in this column for example I sometimes put a note then a dash and the cMs |
| Programming is how I make my living so a small donation of $5 or $10 via Paypal would be most appreciated, if you find this tool useful. |
7 Comments
I got a question about this mapper via email which is a question that I see often in different forms so I thought I would answer it here. Eric asked:
To which I responded:
The current technology cannot separate which half so the only way to tell is with triangulation techniques
My one chromosome mapper just makes a picture from the data you provide
The only way you can tell is by comparing the people you match on a segment to each other which you can do at GEDmatch and 23andme (if you are sharing) but cannot do at family treeDNA. However if you have more than one test to work with you can sometimes figure it out
Try this post
http://blog.kittycooper.com/2014/06/another-way-to-triangulate-using-close-relatives/
Hi
Using this one Chr mapper, what does it mean if I have 22 people sitting directly on top of each other in the same spot?
and
in you other mapper, does a person who appears on more than one Chr mean a more legitimate association with oneself?
and
I get no maternal and paternal seperation results, even though my son has a 23andme file and has a split view screen for paternal and maternal results.
kind regards victor
Victor –
There is a mailing list at Yahoo called DNA-NEWBIE which would be a great place to pose some of these questions. Here are my answers.
1) My mapper is just an image of your data. If you have 22 people in one spot, either you had a prolific ancestor whose DNA has passed to many or this could be a “Pile up,” something I intend to blog about soon, which is discussed in this article http://www.isogg.org/wiki/Identical_By_Descent#Excess_IBD_sharing
2) a person whom you match more than once for good-sized segments (both greater than 7cM roughly) ether shares more than one ancestor with you or is more closely related (3rd-4th cousin) unless you are both descended from an endogamous group like Ashkenazi, Mennonites, or Polynesians. The size of the segment is also a good indicator, the bigger the better.
3) to get maternal and paternal split views you must have tested one of your parents. Your son has you.
Thanks
I am a newbie……I do not use yahoo, i will try to find DNA-NEWBIE
on 1 – looking forward to your article.
on 2 – zero on endogamous group , unless 0.1% ashkenazi still counts
regards
victor
A question that I cannot find the answer for is this. On one of my chromosomes, only on one, I have a stretch of all yellow with a few green bars at Gedmatch. I have great variations in this with my matches. Always on my mother’s side, my matches, not all but a good number, have this abnormal looking variation, some much larger than mine.
When I run this at Gedmatch the bar at the bottom does not show blue ( a section matching) However, when run on a triangulation it is a definate match on this area. Then if I run it one to one it shows no match. This is very troubling to me.
Thank you for considering this.
Evelyn, you only get the blue line with a match of 7cM or more so it sounds like your matches for that area are smaller.
See my post about using GEDmatch http://blog.kittycooper.com/2014/10/using-gedmatch-for-my-ancestry-com-cousins/
When you have many smaller matches for a specific segment they can be what is called a “pile up” or a bit of DNA from an ancestor too far back to find that you share with many many folk. A blog post to be done soon …
an organism’s genetic material is located where?