When I first started doing DNA testing and I saw that full siblings shared only about 2800 cM of their DNA, I was surprised. I expected more. Then I thought about it. We have 23 pairs of chromosomes but the testing process cannot separate the two parts of each pair so our matches are seen as if we had only 23 unpaired chromosomes.
The matching segments are listed and totaled as if there was just one side of each pair even though it is using the data from both sides for the comparison; that is why they are called half identical regions or HIRs. Click here for my blog post with a deeper discussion of why doing comparisons that way can create false matching segments.
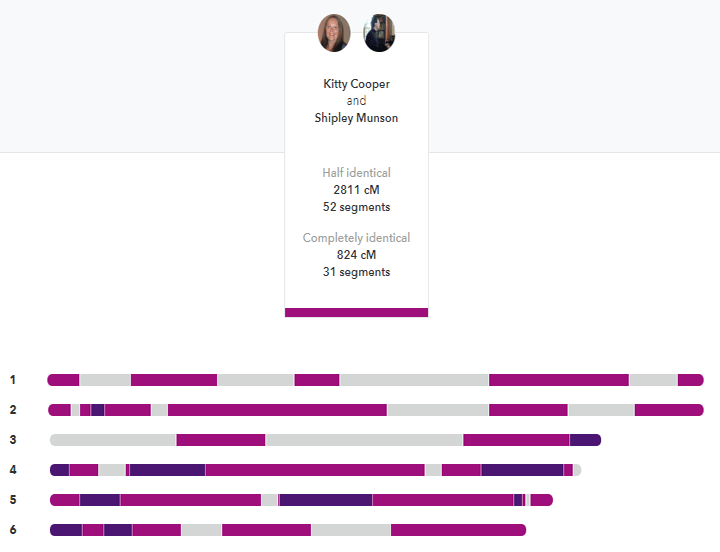
My brother and I compared at 23andme. The darker segments are the FIRs. Click it for the full image.
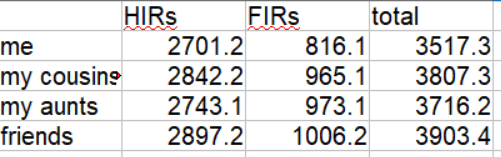
Siblings are usually listed as sharing about 50% of their DNA but that is only the half identical part that most companies measure. How many places did they get the same DNA from both parents on both pairs of a chromosome? In other words how many fully identical regions (FIRs) do siblings share? In my experience, they share about 800 cM or so that are fully identical. Thus the total of the FIRs and HIRs is about 3500-3700 cM; the same amount expected of a parent match. That intuitively makes much more sense to me.
So what would be the logical basis for this? Each parent gives each child half of their DNA but not all of it will be the same half. Roughly half of what each parent gives me will be the same as what my brother got so about 2 times 1800 for 3600; yet most full siblings are shown as sharing only about 2600-2800 cM of half identical segments. The rest of that 3600 is found in the fully identical segments.
A recent query I got was “I only share 2553 cMs with my sibling are we full or half?” By the online calculator at DNApainter they are full siblings sharing a low amount, but another possibility is the father of one sibling was the brother of the other. That is called ¾ sibings. To tell the difference total up the FIRs and the HIRs. If they total less than 3000 rather than the 3600 for full siblings then they are most likely three quarters siblings or some similar relationship.
23andme totals both the FIRs and the HIRs unlike any other testing company, as shown above, however those totals include the X which throws it off a bit for my calculations.
 Wherever you tested, you can get the FIRs and the HIRs by comparing the two kits at GEDmatch.com.
Wherever you tested, you can get the FIRs and the HIRs by comparing the two kits at GEDmatch.com.
Here’s how. Presuming you have both uploaded to GEDmatch (and hopefully both opted in to helping law enforcement identify violent criminals and victims – click here for my post on that), start with a One-to-One comparison of the two kits.
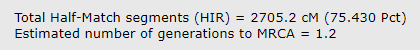
The default comparison after you click submit shows the HIRs (half identical regions) and totals them at the bottom as shown below. The total for my brother and I is 2705.2. Since this does not include the X chromosome it is slightly lower than the total at 23andme.
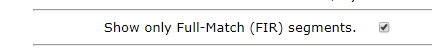
Now click the back button on your browser to return to the form with your two kit numbers and check the box that says “Show only Full-Match (FIR) segments. “ then submit again.
For FIRs my brother and I have 816.1 cM. Adding 816.1 to 2701.2 gets us 3507.3. Now that’s more like it!
Even though this logic seems clear to me, I went ahead and looked at a few more examples of full siblings in my data.
So the answer is to look at the FIRs if you suspect you are ¾ siblings or some other relationship. Click here for an example of how an unusual relationship was determined using the FIRs.
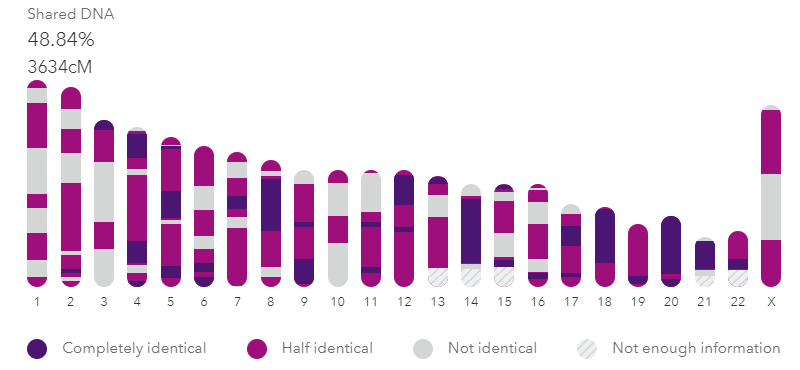
UPDATE 24 Sep 2021: 23andme is now totalling the FIRs and HIRs in its summary of the DNA shared on the match page when you click on View DNA details. Here is how my brother and I look there now
I have a case where two men supposedly are brothers and following your steps above they match HIR 2518.5 and FIR 537.1.
Logic, location, etc. points to the fathers being a father/son combo. One brother shares DNA matches of the son’s mother side of the family and other doesn’t.
Half brothers, 3/4 siblings, ??
My niece (A) who shares 1800cms with me just heard from her birth father(B).. My nieces birth father shares the same birth father (C) with me but my birth mother (D) is a full sibling of (B), so we are 3/4 siblings. Hopefully (B) will do dna testing at FTDNA or ancestry. My birth father (C) was grandfather of (A). Best regards, Doug
I would be interested to see those results if (B) tests
I meant to say I have an example with 2 siblings sharing 3170.1cm (2521.5 and 648.6 fir). Do you think this is an example of what you spoke about in your last post?
Thanks
Bill Hickey
Yes, another way to get 3/4 siblings is when a father and son have children by the same woman.
This is one of your best posts ever. I’ve puzzled over the rather wildly differing DNA matches for half-siblings, from the different testing sites that they used. These were direct tests, not uploads. Your clear and concise explanation was extremely useful. Now I’ll go back and run the GEDmatch utilities as you suggest. Thank you so much!
Thanks distant cousin,
I rewrote it many times trying to make the concepts clear so this is lovely to hear. But did you mean full siblings in your comment? That is where the FIRs come in.
What about when siblings seemingly share too much DNA? Sib 1 shares 2659 HIR with 894 FIR but Sib 2 shares 3002 HIR and a whopping 1145 FIR. According to 23andme that’s a 10% difference between sibs.
I’m sure that endogamy must be to blame somewhere but wouldn’t that mean that both sibs should share excess amounts with me, instead of one less and one more?
Oceania-
Endogamy does not usually play much of a role in close family matches unless the parents are related. While there can be a wide variation due to the randomness of DNAthis seems extreme.
23andme includes the X and segments down to 5cM. Try totalling just the segments 7cM or larger excluding X and see what you get.
Better yet if you all upload to Gedmatch and do the comparison there plus check if your parents are related. And please be a good citizen and opt in to helping law enforcement identify victims and violent criminals
Thanks Kitty. I’ll give that a try. According to Gedmatch my parents aren’t related. I am opted in for law enforcement too.
Thanks for opting in Oceania – now do one to one comparisons there with each sibling as per this article and report back with the new numbers!
Hi Kitty. I have done extensive research trying to discover my paternal grandfather. All the info and DNA matches are totally confusing me. There is a definite link or possible answers, but i am blind to it.
Can i ask for help please?
Hi Annie –
There is a Facebook group called DNA Detectives where you can request a search angel as well as others. See the list of resources here:
https://blog.kittycooper.com/more-dna/help-for-adoptees/
I do help people also but I charge for that I will send you an email.
Technically each of the 23 chromosomes for each parent is passed at a 50% chance of matching and it is possible to win the coin flip all 46 times. So there is no absolute upper limit in sibling dna match, only statistical likely goods. 1/(2^46) is as close to zero as you can get but it’s still possible. That is easy for me…this 2,500 full sibling thing has me searching. It says 2559 is between 45 and 52% shared dna but 7400 is the total
Thank you for your interesting post. I tried the GEDmatch technique and came up with 3221.5cM total (2454.6cM HIR + 766.9cM FIR). Could this be full siblings or does it clearly indicate 3/4?
Full siblings is the most likely with An FIR total so close to 800
Thank you Kitty!
Interesting post. My GEDmatch results are 2442.6 and 550.4, total 2993.0 cM, with basically no possibility of anything but being a full sibling.
Interesting. There is always a variation in DNA results. Your numbers would tend to be a 3/4 but obviously can also be a full sibling.
Do both of you have matches on your father’s mother’s side? If only one of you does, then there is the possibility of the other way a 3/4 sibling occurs: a father and a son having children with the same woman.
Both of us match on our father’s mother’s side.
And the answer is that there were far fewer SNPs used for Fred’s sibling comparison because they were tested on different chips!
At the bottom you can see something like
405732 SNPs used for this comparison.
You want to have some 800K plus to compare, here is mine with my brother
876580 SNPs used for this comparison.
For my results above, I am using 600266 SNPs for this comparison. Does that change my situation? I don’t think so as they still have different matches-one matches son’s mother relations and other doesn’t. Just trying to be sure….
Bill –
600K plus is usually OK. The more important fact is the lack of matches to the one son’s mother’s family
Kitty, thanks for your response. That is what I also believe is the defining element.
Hi Kitty,
I was under the impression that the HIR amounts identified in the one-to-one comparison on GEDMatch are, in fact, the SUM of the HIRs and the FIRs. I recall learning this in the Genealogy Tips and Tricks Facebook group. What am I missing? Thanks!
Chris –
Either you misunderstood or whoever told you that mispoke. That is just not the case.
Look at the screen shots for me and my brother above and see that the it says in the normal one to one “Total Half-Match segments (HIR) = “. Also notice that the number just a tiny bit smaller than 23andme totals …
Hello again,
I’m curious about something else that ties into this (please bear with me, this is still sinking in! 🙂 ). I’m reading/inferring that you mean the sum of the HIR and FIR should be in the 3500-3700 cM range. Per the ISOGG Wiki, full siblings are expected to share roughly 25% full identical regions, 50% half identical regions, and 25% no shared DNA. If you are adding HIR and FIR and getting the total amount, what happened to the 25% that isn’t shared?
The problem comes in the way percentages are used. The norm is to use precentages in reference to just half your DNA since the HIRs are all most DNA testing companies show.
In other words, the statement you are quoting is confusing. I will see if I can get it corrected.
The numbers you are quoting are based on half your DNA, if you look at a diagram. About 25% will be FIRs and and another 25% will be HIRs thus 50%. See this post for the 25% explained: http://www.andreasancestors.com/2011/12/questions-from-my-inbox-siblings.html
In other words this statement later in the article explains it better, “Thus, for example, the shared cM for full-siblings will on average be 75% of the total length of the genome, of which on average 50 percentage points are half-identical and 25 percentage points are fully identical.”
Hi Kitty! I almost forgot about this. I think I get it now. Thanks for your patience! 🙂
I’m so glad that you’re reminding people to opt in to help law enforcement at GedMatch.
Do you know what percentage of GedMatch subscribers have now opted in? I know they defaulted everybody in their database to “opt out” when this topic kind of blew up a year or two ago … of course that meant that anybody who was deceased couldn’t opt back in, so those kits were lost forever as far as law enforcement was concerned. And GedMatch emailed everybody, but a few months after that I heard that only 4% of members had taken the time to opt back in? I hope that new members since then have opted in, I’m pretty sure GedMatch puts that front and center when you join …
Last I heard only 200K had opted in out of over a million
Hi Kitty – I have the following brother and sister comparison from GEDmatch:
Total Full-Match segments (FIR) = 540.0 cM (15.056 Pct)
Total Half-Match segments (HIR) = 2163.4 cM (60.323 Pct)
They both tested on Ancestry. They have same mother but I am suspecting different fathers. The fathers are related as 1st cousins and the fathers have parents who were a set of brothers who married a set of sisters making the fathers double first cousins. I am thinking 3/4 siblings here at 2703.4 Cm, what do you think? Thanks!
I agree with your assessment Lisa
Thank you! I don’t have DNA tests from either potential fathers. Would the GEDmatch X DNA comparison tool be able to help prove my theory any further? Not sure if there is an average match of Cm on the 23rd chromosome for full siblings versus half siblings or if it would be difficult to determine since the potential fathers are related as double first cousins.
If the fathers of the two brothers were from the same parents then I do not see any easy way to prove this. If they were from different parents, so a woman’s brother married her husband’s sister then you could use Y testing on the brother if there is another male child from the 2nd father… Since the brother only has X from his mother, it would not be useful.
I think Gedmatch made the BIG mistake of setting their default to “opt-out.”
FTDNA has their default set to “opt-in.”
Perhaps you might want to turn off the Google Docs feature to “view the results so far” as that exposes the email addresses of the folks who included them.
Very good point William, I removed that feature
My brother and I both tested at 23 and Me
At 23 and Me I get (2678-68X) =2610 HIR, (920-71X) = 849 FIR
At GEDMatch I get 2594 HIR, 811 FIR
In my submission I forgot to subtract the X from the FIR, apologies.
Mark
Mark
Kitty,
I wanted to give you info for your study on my brothers and me but GEDmatch is not showing the total cMs FIR just Pct SNPs.I do see the green FIR segments but no sum. Here’s what I got:
Total Half-Match segments (HIR) 2745.6cM (76.561 Pct)
Estimated number of generations to MRCA = 1.2
57 shared segments found for this comparison.
400656 SNPs used for this comparison.
72.509 Pct SNPs are full identical
To get the sum you have to check the box that says show FIR segments only as shown in the article
Incredible article! I was wondering why I shared less Dna with my sister.
Just a (good) question: 2 full brothers, whose parents are 1st cousins, share more than the regular amount of dna between siblings?
yes they will share extra DNA because in addition to being full brothers they are 2nd cousins (about 229 cM)
Hello
My niece (A) who shares 1800cms with me just heard from her birth father(B).. My nieces birth father shares the same birth father (C) with me but my birth mother (D) is a full sibling of (B), so we are 3/4 siblings.
Wonderful Finntech, have you confirmed this by looking at the FIRs on GEDmatch or 23andme?
If so please fill out my survey at https://docs.google.com/forms/d/e/1FAIpQLSe4Q1pep-I8pYo21JJu7p4T-adZuWnuCirtLVRP50gIR1MfgA/viewform?usp=sf_link
Hello
Family rumors have swirled for years. Looking to move toward closure and putting some things to rest. Mother to Bob and Paula is said to have had a rendevous with reported father Wilbur’s brother Dan. Bob and Paula have tried research but results seem to indicate full siblings. Wilbur and Dan’s family insist Bob is Uncle Dan’s child. Any help would be appreciated.
GED Match
Largest segment = 169.1 cM
Total Half-Match segments (HIR) 2757.4cM (76.909 Pct)
Estimated number of generations to MRCA = 1.2
50 shared segments found for this comparison.
104172 SNPs used for this comparison.
70.243 Pct SNPs are full identical
FIR Results
Largest segment = 75.6 cM
Total Full-Match segments (FIR) = 580.2cM (16.183 Pct)
24 shared segments found for this comparison.
104172 SNPs used for this comparison.
70.243 Pct SNPs are full identical
GED file is comparison between Bob and Paula.
Those numbers indicate full siblings. …
Thank you
Checking in Kitty after providing the two kit numbers as requested. Any luck on the deeper dive?
I’ve found evidence suggesting three fourths siblings.
Sorry, busy time of year here, no I did not find anything more. Email me what you have learned, I can look again in a week.
hello, would it be possible to write to you? I have pretty similar results to you and I would like to exchange ideas with you
Sure about a week after Labor Day, on an extended vacation here … will email you privately
Hello,
My mother 3441cm my sister 2823 my around 2600 my niece 1969, but another sister no DNA but 2 daugher 900cm 2 different father.
My question is my sister no DNA could she have different father.
Thank you
Yes she could, that is too low a cM match for full nieces. Look at your shared matches. Do they share any on your father’s side?
Family scenario:
Woman has 3 daughters: A B C
Daughter A has 1 daughter: D
Results conclude that same man (deceased) is the father of daughters B, C, D.
One man has children with mother + daughter.
Aunt/Niece + 3/4 Sisters ??
Cousins + 3/4 Niece/Nephew ??
Comparison of daughters B + D (43 years as aunt/niece):
Relationship – Sister
Share all ancestors, but each inherited a different mix of their DNA.
Shared DNA
40.00%
2976cM
Comparison of male (son of daughter C) + daughter D
(30 years as cousins):
Predicted relationship
Nephew/Aunt
Share DNA that was passed down from her parents (his grandparents).
Shared DNA
22.67%
1687cM
It would be interesting to see the FIRs for B and D who are both paternal half sisters and maternal aunt/niece so roughly 3/4 sisters
I am a volunteer “Search Angel” & would love some help! The adoptee is 68 & just found out who her birth mother was. I did a “mirror tree” based on her mother & her very close DNA matches from AncestryDNA. I speculated that her birth father was the husband of her mother’s sister & found a possible grandniece who shares 842 cM’s & 36 segments.
Then the adoptee did a 23andMe.com test & found a match to the 79 yr old son of the man who I had speculated was the birth father of the adoptee.
Is the match high enough to be a 3/4 brother? Could they be double first cousins instead?
The match is:
1956cM
Half Identical – 1742cM, 39 segments
Completely Identical – 215cM, 10 segments
mtDNA-the same
NO shared X
Good work Joy! Sorry for the late reply. I have been very busy with other things… Yes your theory works with those numbers.
Thank you Kitty! Do you mean that my original theory of 3/4 brother works?
Yes
Hello Kitty,
Here is what my brother and I get on Gedmatch (raw data comes from FTDNA):
With HIR:
Largest segment = 131.7 cM
Total Half-Match segments (HIR) 2846.8cM (79.371 Pct)
Estimated number of generations to MRCA = 1.2
69 shared segments found for this comparison.
452336 SNPs used for this comparison.
81.645 Pct SNPs are full identical
With FIR:
Largest segment = 31.2 cM
Total Full-Match segments (FIR) = 597.7cM (16.664 Pct)
46 shared segments found for this comparison.
452336 SNPs used for this comparison.
81.645 Pct SNPs are full identical
Are the results above normal for full siblings? Last but not least, is the largest FIR segment at “only” 31.2 centimorgans usual?
Cheers,
Best Regards!
Normal range, no worries!
Hello, what about:
HIR:2505 CM
Largest segment: 146 CM
FIR: 616 CM
L.S.: 69 CM
TOTAL HIR + FIR= 3121 CM
????
still above 3000 so probably full siblings
Hi Kitty, I found your blog after watching the most recent Youtube video by Family History Fanatics (they discuss some interesting features of chromosome browsers and mention your post on using the tool to unveil quarter relationships).
I was curious to know whether there’s any way to solve family mysteries given segment data from relatively more distant relatives. In my case, I want to discover who was my grandfather’s father (there are two known possibilities). After testing him in multiple companies, I found one granddaughter of my grandfather’s eldest sister. They share about 507cM on Gedmatch, and the most extended segment is about 50cM (these numbers change slightly depending on the company, but the range goes from 480-520cM).
One possibility is that they are entirely related (e.g., she’s my grandfather’s great-niece); another is that my grandfather and his eldest sister were only half-siblings. I don’t have anyone closer to test; he’s the youngest of 12 siblings and the only one alive. His father passed away when he was a kid, and his mom married again, presumably with his biological father. DNA Painter suggests there’s a 90% chance he and his great-niece are half-related and 10% entirely related. Since there’s an overlap between these scenarios, I’m looking for additional evidence. However, he has no matches above 40cM that I can safely attribute to his father.
Do you know any research trying to establish genetic/genealogical proximity using DNA segment size or anything? Thank you very much.
Sergio,
A Y test at family tree DNA could solve this. If he had that many siblings surely you can surely find a son of one of his brothers to do a Y test also. That would be conclusive as to whether he had a different father.
Read this blog post of mine about a similar mystery which may help you
https://blog.kittycooper.com/2020/05/who-is-my-great-great-grandfathers-daddy-a-thrulines-experiment/
Kitty, this is excellent; thank you so much for the suggestion. My grandpa’s official father was from northern Portugal, and the presumed dad was from northern Italy, so I think the chances are very high that I would find a different result in a Y test if my grandpa’s brothers didn’t share the same father. I just need to find these relatives.
Hello,
I have a question about a sibling relationship where I don’t know whether I should classify it as a 3/4 sibling or as a full sibling.
On gedmatch, the two share 2741cM (HIR) and only 593 (FIR) and come to a total of 3334cM. what is your assessment?
here are the results of gedmatch in detail:
Largest segment = 132 cM
Total Half-Match segments (HIR) 2737.9cM (76.394 Pct)
Estimated number of generations to MRCA = 1.2
69 shared segments found for this comparison.
196510 SNPs used for this comparison.
68.094 Pct SNPs are full identical
Largest segment = 49.1 cM
Total Full-Match segments (FIR) = 594cM (16.573 Pct)
25 shared segments found for this comparison.
196510 SNPs used for this comparison.
68.094 Pct SNPs are full identical
That is slightly low for full siblings but still quite possible. 3/4 is also likely. Do you have other evidence? Family stories? Y? Sorry I cannot be more definitive.
Hello Kitty,
well, the two subjects have a half-sister. a female paternal cousin confirms that the paternal grandparents must be the same in both subjects. that means on this side at most a full brother of the father would be possible. both also share the same haplogroup. paternal and maternal. According to the theory of genealogy, the only question left is that one is not the half-sister but the aunt, which could only mean that the father of the two “brothers” had a child with another half-sister on the mother’s side. However, nothing is known about this and it would be rather unlikely since the mother and the resulting half-sister had to have a child at the age of 14. you see, it’s a bit of a tricky situation…
The numbers do suggest full siblings. There are a number of online simulations you can look at the charts here
https://isogg.org/wiki/Three-quarter_sibling
and this article
https://dna-sci.com/2021/08/05/3-4-siblings-you-can-tell-the-difference/
Hello kitty, I was able to do some more research. In the meantime I was able to test a cousin from my father’s line. She is the daughter of another of my father’s brothers. Our shared DNA was 585cM, which unfortunately for me is second proof of a 3/4 sibling relationship between my father and his brothers.
Hello Kitty
sorry if I bother you again with the same topic. I was able to test another first-degree cousin, with whom I had an agreement of 930cm. He is the brother of the cousin with whom I only had a match of 585cm. I think this would still be a strong indication that the fathers were full siblings, am I interpreting this correctly?
Dave
Sorry for the delay getting back to you. Yes 930 is a nice number for a full first cousin, thus fathers being full brothers
Hello Kitty
I hope I can ask you something about this topic again.
I have been able to do some tests in the meantime.
I would be interested in your opinion on this.
As mentioned above, the results of the two brothers tested are in the range of a full sibling, but based on the amount of FIR (634cM) they could also belong to a 3/4 sibling (HIR 2770cM; IBD 3400cM).
To investigate this further, I was able to test two of my cousins and one female cousin (children of my father’s brother). Two of the cousins were in the plausible range of 880-930cM, one female cousin was at 630cM. The cousins had a similarity of 1760-1850cM to my father. Overall, I would rate them as full siblings, the only thing that confuses me is the low value for my female cousin. the remaining values then correspond to the normal range again.
thank you for a short feedback
DNA inheritance can be quite variable, the more so the further away the relationship. If you click on the box for 1st cousin at the link below, a bar chart of frequencies will appear and you will see that that lower match is within normal range
https://dnapainter.com/tools/sharedcmv4
What a great article. Will I be able to use FIRs and HIRs of a match compared to someone I am helping to determine if the match’s father is a 3/4 sibling or full sibling to the person? The match shares 1506cMs and we haven’t migrated to Gedmatch yet.
Thank you.
You would need the father’s DNA. The child gets half their DNA from each parent so no FIRs with the match unless the mother is also related to the match.
However that amount of cM is almost surely a full niece/nephew as per this calculator https://dnapainter.com/tools/sharedcmv4
You might also try this calculator
https://dna-sci.com/tools/segcm/
Bill if that is a first cousin then that amount is on the low side so does prove anything nor change the 3/4 likelihood