I suddenly realized that I could find the people who match my late husband Steven M Cooper on his mutated section of the BRCA2 gene at the various DNA testing sites that show chromosome information. His particular BRCA2 mutation, implicated in breast cancer and melanoma, likely contributed to the fatal outcome of his prostate cancer. A problem with doing this is that none of his tested family members match that gene, so there would be no way to know if his matches had his bad maternal BRCA2 or his good paternal one.
So should I contact those many people and warn them? I had previously alerted his known LILIEN side cousins to this issue and already a second cousin once removed on his LILIEN line discovered her breast cancer early due to my warning. However I may be sending a false alarm to many. I decided it was something I should do. My family history investigations left me confident that Steve’s mutation came from one of the the parents of his grandmother Amalie LILIEN Tieger (jewish) born in Kalusz, Ukraine. Of course, it may well have originated further back.
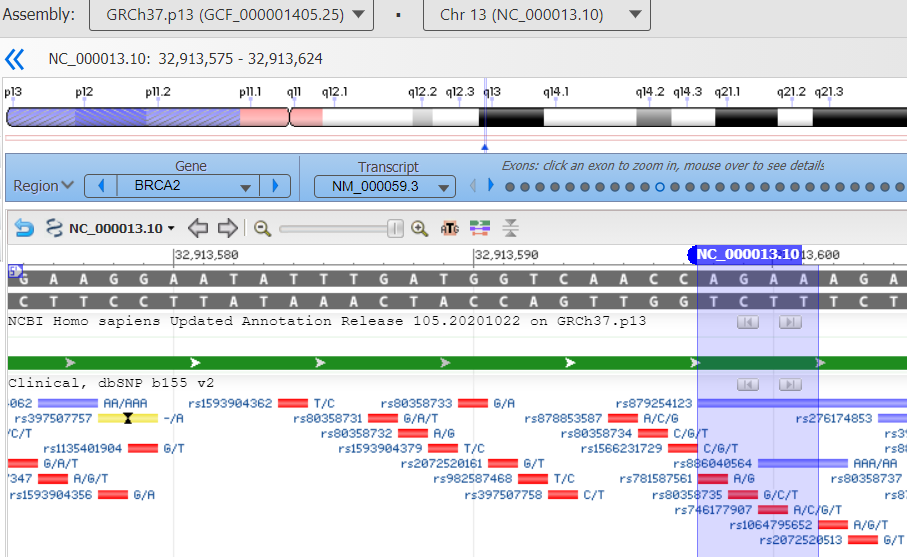
First I had to locate the mutation in a numbering system that would translate to what we get from our DNA testing companies. From various google searches I learned that BRCA2 is located on chromosone 13. Looking at the report from Color Genomics, the test his oncologist ordered, I could see the location was a deletion at base pairs 32,913,602_32,913,605. More googling found that this is not the common Jewish BRCA2 mutation. That explains why his initial 23andme test years ago did not find it. Next I found the actual National Health Institute fact sheet for his mutation (click here) which had a click point to the diagram below. Click here for the cancer.gov discussion of BRCA2
Now to find the people who match him on that segment. I downloaded the full list of his matches with segment information from each site. The easiest site to use was GEDmatch because I could use the segment search function to get just the matches to his BRCA2 section of chromsome 13. At the other sites I had to get the full list of all matching segments and then sort by chromosome and start point to find the matching people.
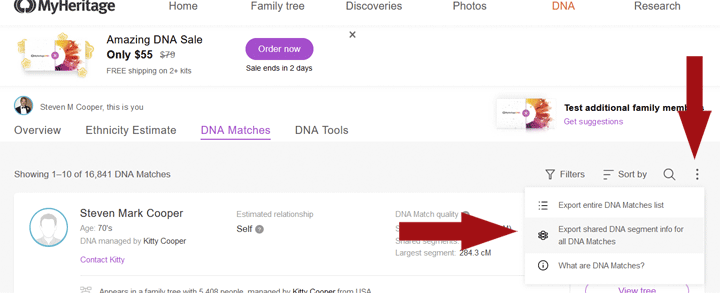
At MyHeritage on his matches page I clicked on the three dots and then selected the export as shown in the image above. At 23andme I used the “Request DNA Relatives Data Download” button located at the bottom of the page that has a list of all his DNA relatives. At Family Tree DNA I went to the Family Finder Matches page and on the top right under the search box clicked on “export CSV.” Ancestry does not provide chromosome data; you have to export those results elsewhere to get them.
By the way there were 9 kits at GEDmatch, 9 at 23andme, 59 at MyHeritage, and 61 at Family Tree DNA, that matched him on the BRCA2 gene location in question. So far I have contacted the Gedmatch and 23andme matches and am about 1/3 of the way down the Family Tree DNA list. It may take me weeks! So far I have heard back from only four of the 25 or so I have contacted.
The main reason I wrote this post is to give those people who match Steve at the BRCA2 location more information about the issue. Perhaps this article will also help others track down genes of interest and the testers that share them.
Nice and informative post!
Genes with clinical relevance are nicely summarized at OMIM: https://omim.org/. The data is open source and usable in my ways. It includes specific location data (chr, start position) for many of the genes. I’ve used it to demonstrate seriously bad genes at pill up regions identified by genetic genealogists. This makes sense because evolution would favor conserving the good genes.
The analytics can be facilitated by use of a graph database.
Very good post! My maternal line is rampant with breast cancer and ovarian cancer. We can document those cancers as the cause of death in every woman in our maternal line but one -my maternal Grandmother- back through to my great-great grandmothers. Three of my five sisters have the mutated BRCA2 gene and have had breast cancer, including the sister we never knew who was adopted. I was able to locate her and inform her of the family history which, in turn, caused her to find the cancer early. Thank you for the informative post on how to locate the gene in our tests.
Without having a known person with the mutation at that location, how do I examine the raw data file data for the person’s chr 13 segment to determine if the mutation was present? I can identify matches on that segment as you describe for a relative I suspect may have the gene.
Also, would it be possible to use DNAPainter to identify the matches at that segment and create a group of those matches?
If you don’t have a known genetic problem to resesrch then perhaps what you want is to find the health results in your DNA test with a 3rd oarty web site like Promethease see https://blog.kittycooper.com/2019/12/free-health-results-from-your-dna-test-for-the-rest-of-2019/
Without a tested relative that is known to also have the mutation DNApainter cannot help
So sorry about your loss. This is very interesting to me. Not only did my husband have an early prostate cancer (unrelated to BRCA) but our son-in-law is an oncologist with a degree in genetics, who specializes in prostate cancer research, particularly late stage. A little over a wk ago, he told me of his new publication re a difference between BRCA1 & BRCA2 in prostate cancer. I will try to forward your information to him.
Also in unrelated research, we discovered in the last yr a rare mutation in my family on Ch.7. A study is being conducted on us by Johns Hopkins in which they are drawing our blood to look at the genetics. I decided to see if I could discern anything by comparing our DNA in the genetic position of the mutation for those of us already tested across genealogy DNA sites. This included siblings & two subsequent generations to a more limited extent, as well as transferring some to My Heritage to use their chromosome browser. Out of the 15 I looked at, there appears to be only one incorrect prediction of the mutation when compared to the results of their study, probably because of the transfer, different chips or incomplete testing of markers on the genealogy sites. I hope to find the time to look at this further on Gedmatch. I am also looking at an uncle & cousins. In my search for the mutation founder ethnicity, it appears that the mutation match is only with cousins in Norway (my paternal great-grandfather was a Norwegian emigrant) & when I messaged them, two of the three that responded had an aunt who had either melanoma or a specific brain tumor, symptoms of the mutation. Time might tell whether this has any validity to my hypotheses.
Ann
I too have a genetic disease and have always been on the lookout for people who share that segment. I have only checked on gedmatch but you are inspiring me to check the other sites. In my case it is a rare single nucleotide mutation that originated in County Donegal, Ireland. The disease is devastating and life threatening, It is called amyloid polyneuropathy. I found out I had it about 10 yrs ago when I did 23 and me. Thankfully they came up with a mRNA inhibitor treatment about 5 yrs ago that for me actually reversed the disease process. I never would have known about the treatment if I didn’t know whatI had. So thank you for your post. I was somewhat hesitant to contact strangers with this bad news. But I do know I inherited it from my father and have my mother.’s dna, so I can be sure that anyone who matches me there has at least a good chance of having the disease. As a side note I was able to test my sibs, cousins and kids on 23 and me without buying the health package. You can just look it up in an easy to use mutation search on their website.
The BRCA2 mutation you mention is on newer 23andMe chips, where it is reported under the name i704424. The normal genotype is reported as “II”, and someone carrying the mutation will be “DI”.
In general, though, people should be aware that that there are over 3,000 known BRCA1 and BRCA2 mutations, not to mention mutations in other genes, which significantly increase the chance of developing breast, ovarian or other cancers. Instead of using DNA chips (regardless of the company), which actually lack most of the known mutations, the most thorough way to test is via DNA sequencing. For those with a family history of cancer, there are dedicated gene panel tests that completely sequence just about every relevant gene. One list of companies offering such tests is on the FORCE webpage, https://www.facingourrisk.org/info/hereditary-cancer-and-genetic-testing/how-to-get-testing/where-to-get-testing.
There is an rsid for the mutation found in Steve, rs879254123. 23andMe does have coverage of this position on the v5 chip with one of their custom probes, i704424.
If Steve didn’t test on v5, I’d be curious to know if he might have had a no-call in the vicinity of 32,913,602. The deletion of 4 bases could conceivably interfere with probes for nearby SNPs.
The fact that 23andMe included the mutation in their allotment of custom SNPs would seem to imply that it’s common enough to be worth including — and therefore that the mutation probably originated further back than your known ancestor. I did find a reference to the SNP in a small Greek database, so that could also imply it has been around for a while.
http://ithaka.rrp.demokritos.gr/CanVaS/view/BRCA2?search_VariantOnGenome%2FDBID=%3D%22BRCA2_000270%22
Steve’s 23andme test was v3. I will look at it again.
Interesting. The V3 chip definitely does not show this. His BRCA2 listing skips from 32913562 to 32913770 the only no call is a bit further on at
BRCA2 rs34309943 32914712
I checked a cousin who has a v4 test and that SNP – rs56087561 32913562 A or C is in fact tested in v4. Plus it has a click point to the NIH report here https://www.ncbi.nlm.nih.gov/snp/rs56087561?vertical_tab=true
Just to clarify, the SNP for Steve’s mutation is not included in v4. The one you looked up is at a different position.
Readers of your blog might also be interested in my experience for the opposite scenario. You have a known mutation and are looking for cousins who might share it. I have known cousins with hearing impairment, and we were looking for the mutation.
http://blog.23andme.com/23andme-and-you/23andme-how-to/tracking-down-a-trait/
http://blog.23andme.com/23andme-customer-stories/citizen-scientist/
Good point. My son and father both have an unusual mild blue/green color blindness issue which must be on the X since I do not have it. last I looked at the literature they thought it is on chromosome 7 . I found other family members with it .. So next project. Is to place it.. … http://kittymunson.com/index.php?page=colorblindness
I didn’t realize melanoma was connected to this gene. I’ve had two removed and it came up #1 on Promethease report. I need to dig deeper. Thanks for the info.
I commented on this previously. I want to add that the mutation my family carries is also related to melanoma but not to BRCA. Further, the specific genomic position, or range, & chromosome of our mutation was easily found by looking it up on Wikipedia.