There is a wonderful site at http://www.y-str.org with many good tools for Y DNA and autosomal DNA that run as programs on your PC plus a cool ISOGG Y tree add-on for the Chrome browser. My specific interest at the moment is figuring out which Y SNPs are already tested by 23andme so as not to test them again at FamilyTreeDNA since my Dad has kits at both places. I blogged about how to do that manually back in February, but now there is a program that will do that for you. However it took me a while to figure out how to do what I wanted from the instructions given, so I will do a step-by-step tutorial in this post in order to remember what I did.
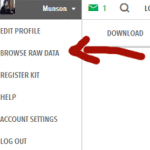 First download your raw data from 23andme by going to the “Browse Raw Data” Page which is listed in the menu that appears under your name on the top right. Then on the raw data page click on “Download” in the second top bar on the right hand side. This takes you to a page with various warnings and requires that you reenter your password as well as the answer to your secret question before it starts the download.
First download your raw data from 23andme by going to the “Browse Raw Data” Page which is listed in the menu that appears under your name on the top right. Then on the raw data page click on “Download” in the second top bar on the right hand side. This takes you to a page with various warnings and requires that you reenter your password as well as the answer to your secret question before it starts the download.
Save the download file somewhere that makes sense for you. I have a folder called RawData in the folder DNA that I use. Once the raw data file is downloaded, you will need to unzip it before using it with the various tools. To unzip in windows all you need to do is open a file explorer window (a manila envelope is the icon) and then right click on the file name to get a little menu that includes “extract all” which is the one to click.
 Now go to http://www.y-str.org/2014/04/23andme-to-ysnps.html and download the program which turns the Y SNPs from your 23andme raw data file into the names in ISOGG format.
Now go to http://www.y-str.org/2014/04/23andme-to-ysnps.html and download the program which turns the Y SNPs from your 23andme raw data file into the names in ISOGG format.
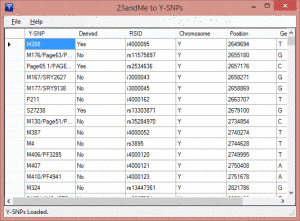
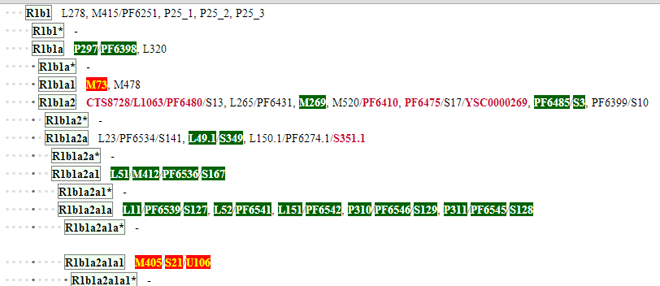
When you run this program on your PC you get a little window. Click on “File” in the upper left and then the menu item “Open 23andme raw-data.” Use the file box that comes up to find the unzipped text file from your raw data download. Then it will translate the data into ISOGG named SNPs as in the image on the left. Now click on “File” again and then “Save Y-SNPS.”
Now you have a text file of the SNPs which you can use with the chrome add-on to see where you fall in the current ISOGG tree. This new file can be opened with notepad or wordpad. Click on it in the file explorer in order to open it and have the data handy for the next step.
To get the chrome add-on go to this page http://www.y-str.org/2014/04/isogg-y-tree-addon-for-google-chrome.html and click on the words in red “Install: ISOGG AddOn Chrome AddOn” (or click it here).
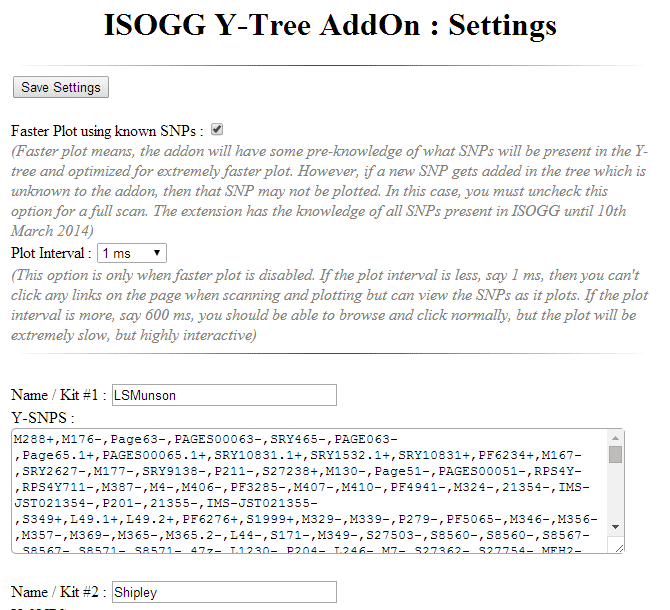
Once you have installed the ISOGG add-on, you need to put the Y SNP data you created above into the options for the add on. To do that you need to click on the three red lines at the far top right of your chrome browser and then in the resulting menu click “Tools” and then “Extensions.” This will show a list of your extensions, the ISOGG one looks like this:
Click on the blue “Options” to get to the page where you input the Y SNP data. You can input the data from up to 10 kits there. Using the edit menu in your notepad that has the Y SNP data open you can select all and then copy all in order to paste it into the box for your kit on the options page for your add-on.
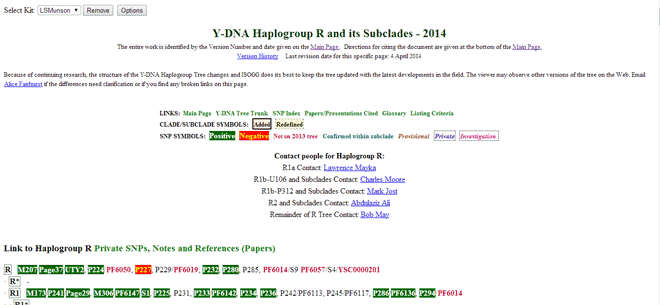
Once you have the Y SNP data loaded, you can go to your section of the ISOGG Y tree to see your tested SNPs highlighted. The URL is http://www.isogg.org/tree/ then click on the letter for your haplogroup, for us it is R. On the page for your haplogroup you will see lots of new stuff. First there is a select kit dropdown box on the top left. Then every SNP that has been tested by 23andme is shown in green if it is positive and orange if it is negative. Note the options button, this is a quicker way to get to the options page for the ISOGG add-on.
In our case the SNPs indicate R1b1a2a1a
Next to look at my brother…
Thanks to user Axel on 23andme, who pointed out a thread about these tools in the community there.

Also there is a good step by step on downloading your raw data from 23andme here: http://www.haplogroup.org/23andme-raw-data-file-download/
Great instructions. Great utility! Very Informative
Here are the Geno2 R1b SNPs as I understand them. This txt file also worked in the extension after the line breaks were removed.
CTS8728+, L23+, YSC0000072+, L51+, L151+, L21+, DF21+, DF25+, CTS3655+, L627+, L625+, CTS1970+, PF4252+, DF23+, M222+, PF3292+, PF3988+, F3952+, F3024+, CTS8002+, M226+, F499+, Z70+, CTS12173+, PF2028+, PF1909+, CTS8580+, CTS3771+, CTS10488+, F1400+, CTS9501+, PF910+, PF7301+, F3637+, CTS6+, F1636+, CTS11548+, F1732+, F1033+, PF4837+, Z255+, Z253+, F3808+,CTS1202+, F1969+, L554+, L226+, CTS11722+, PF5236+, PF7379+, CTS2501+, L580+, L513+, CTS3087+, CTS6919+, F4006+, L371+, CTS10334+, CTS4466+, L270+, M481+, CTS11459+, CTS2457+, PAGES00072+, L96+, M18+, L194+, Z195+, SRY2627+, Z202+, Z205+, Z207+, CTS4299+, CTS8234+, F3198+, F1287+, CTS10893+, PF1023+, L163+, CTS606+, CTS5284+, CTS7416+, F2536+, PF2462+, CTS9454+, CTS4015+, L165+, L247+, PF4354+, PF4280+, CTS4188+, CTS3437+, F3161+, PF6913+, Z274+, Z210+, Z295+, CTS12074+, Z214+, M153+, CTS10504+, PF2462+, F2735+, CTS7927+, CTS4065+, CTS7768+, U152+, L2+, Z49+, Z150+, s42+, CTS7970+, CTS8125+, CTS11381+, L552+, CTS1789+, Z51+, L562+, Z57+, Z148+, Z149+, CTS11232+, CTS278+, CTS4521+, F3799+, F1947+, PF27+, F1036+, P329+, P37+, M75+, F2083+, F1993+, CTS3409+, Z367+, L20+, L739+, F1948+, F1398+, L144+, CTS6911+, F2421+, CTS8178+, CTS9044+, PF7623+, CTS1640+, CTS3936+, CTS5481+, M126+, L196+, L135+, PF7610+, CTS7010+, CTS5153+, F3916+, PF6610+, CTS10009+, CTS3080+, CTS667+, CTS7491+, CTS5689+, CTS1166+, F4205+, PF4367+, CTS6942+, F1643+, Z192+, PF6652+, CTS11874+, CTS8137+, L466+, PF6653+, PF6659+, CTS11022+, CTS6975+, Z36+, L671+, P61+, CTS5531+, CTS1595+, F3372+, Z37+, Z38+, PF257+, CTS10641+, CTS319+, Z191+, CTS188+, CTS4333+, CTS7983+, CTS8516+, CTS2687+, CTS7618+, Z145+, Z146+, CTS2376+, CTS12058+, F2229+, Z72+, CTS7193+, Z46+, Z48+, CTS8949+, CTS6452+, CTS7325+, CTS5478+, CTS10855+, PF4363+, CTS2752+, L4+, CTS5029+, CTS1034+, Z225+, CTS234+, DF19+, CTS11567+, F2691+, CTS10429+, U106+, Z381+, Z28+, CTS2509+, Z2+, YSC0000275+, Z11+, L148+, L47+, L44+, L163+, L46+, L45+, L493+, PF3188+, CTS5603+, Z350+, Z160+, CTS6353+, F2513+, Z191+, CTS603+, U198+, CTS9715+, F1881+, Z156+, P89+, M93+, L1+, M323+, Z14+, L257+, CTS4528+, PF6714+, CTS6889+, PF3252+, P53+, CTS7822+, CTS9219+, CTS11767+, L25+, CTS9940+, PF3928+, F2610+, CTS1039+, CTS2791+, CTS8563+, CTS699+, CTS7763+, CTS1848+, F2107+, Z1862+, CTS6937+, M64+, F2863+,
Once you have a text file of the SNPs you can also paste them into http://ytree.morleydna.com/ instead of having to install the Chrome add-on. It uses the 2013 ISOGG tree but 23andme SNPs markers are older than that anyway so it will still provide you with the 23andme terminal SNP.
I was under the impression that the result of using this utility would be a description of the haplotype – eg E1b1b1b2a, or E1a2b, etc. Am I not seeing this output because the utility does not do this or is another step(s) required?
And also in the textfile there exist data such as page0063, page0078 – should these first be deleted? And also – can one input the entire genome data or should only the Y data be used?
The utility just gets you the SNP names from the full raw data file downloaded from 23andme as described above. The haplogroup can then be found using the chrome addon with the ISOGG page, again explained in this post. Also some of the comments here detail other uses for the SNP list
For haplogroup descriptions see http://www.eupedia.com/europe/origins_haplogroups_europe.shtml
Thanks – I got the link to this page from Alex, and it is very useful. Worked like a charm for the first time. – Doug@Austin
Thank you for this info, I got my basic Ftdna-haplo much deeper with this result from my 23ndme-result.
Easier now continue the search.
The 23andMe To YSNPs tool is now at http://www.y-str.org/2014/04/23andme-to-ysnps.html
Thanks Armando, somehow a space had crept into the URL perhaps on a recent edit, now fixed.
Armando also wrote a detailed reply on this topic, terminal SNPs for us R1bs at
http://archiver.rootsweb.ancestry.com/th/read/GENEALOGY-DNA/2014-04/1398097398
Recommended reading for R1b folk
Armando can also be found at the Yahoo group for us R1bs
https://groups.yahoo.com/neo/groups/R1b-YDNA/info
This is great! Thank you.
Sending much Thanks for CHANCE of me coming across this. VERY Helpful!!!!
Dropped close to $300 at FTDNA, to know I’m M269 and awaiting SNPs that you just showed me how to find with 23andme data. . .Well, I did want to be in their Projects. (I see why FTDNA stopped taking 23andme uploads after 2013, to make cash).
Thank you!!
Please don’t blame family tree DNA, 23andme was the one who changed their chip and made it incompatible with the other testing sites
As to the SNPs, yes my project managers kept asking me to test my R269 dad … I did eventually test p312 which 23andme did not test, we are positive .. But it really is a question of your goals, we Y tested to prove a postulated line of descent for which SNPs where not important but if you want to know your deep ancestry SNPs are the thing.
See this article from Roberta Estes on SNPs v STRs
http://dna-explained.com/2014/02/10/strs-vs-snps-multiple-dna-personalities/
I just ran this tool from the 23andme raw data for my husband and it highlights almost all of the r subgroups in green so I really can’t tell what he is! They revised the names of the haplogroups so now he’s R-M412 instead of R1 something or another. We’re trying to find his family line from the Yoder DNA project and this made it very difficult. :/ Any ideas what I can do?
Well I googled R-M412 and found this wikipedia article which has a nice easy to understand chart and says this used to be R1b1a2a1a*
https://en.wikipedia.org/wiki/Haplogroup_R1b
As to all the groups turning green, if they should not be then try writing to the author of the tool
I have installed the Chrome plug-in, entered my SNPs from 23&me & activated the plug-in, but unfortunately when I go to the ISOGG page for haplogroup R (https://isogg.org/tree/ISOGG_HapgrpR.html) the plug-in does absolutely nothing. No plug-in menu with kit selection & options & no high-lighting of SNPs. Have disabled all blockers et.c. but nothing works. Have ISOGG changed their pages causing the plug-in failure? Grateful for any hints!
Same is happening with me if it makes you feel any better. I have yet to find a solution.
It is possible that it is broken. You can get your Y haplogroup by using Promethease see http://www.geneticgenealogist.net/2016/01/how-to-get-ydna-haplogroup-from.html
Hello! If anyone has a resource on how to actually run the program from this page: https://fiidau.github.io/ it would be so helpful! I am so stuck and I can not seem to get the program to open. I also have a Mac, not sure if that is making anything more difficult. Thank you!
Sorry Michelle, not something I can help with. I suggest you find a young person with programming skills to help