The same question seems to come up over and over again among those new to autosomal DNA testing. If I match A and B on the same segment why is that not enough to prove they match each other and we have a common ancestor?
The reason the ancestor is not proven is that you have two strands of DNA on each chromosome (remember there are 23 pairs of chromosomes) and the testing mechanism cannot differentiate between the two of them. So A could match the piece from your mother and B could match the piece from your father or one of them could even be a false match to a mix of alleles from both parents (see my post on IBC for more on that concept)
The way to prove the common ancestor is to see if A and B match each other in the same place that they match you. This is what we call triangulation.
About a year ago I blogged about how, after many years, a change in spelling on the paper trail had led fellow genealogist Dennis to think his wife Kristine was perhaps descended from my great-grandmother’s brother Carl. To prove this I suggested he test her autosomal DNA.
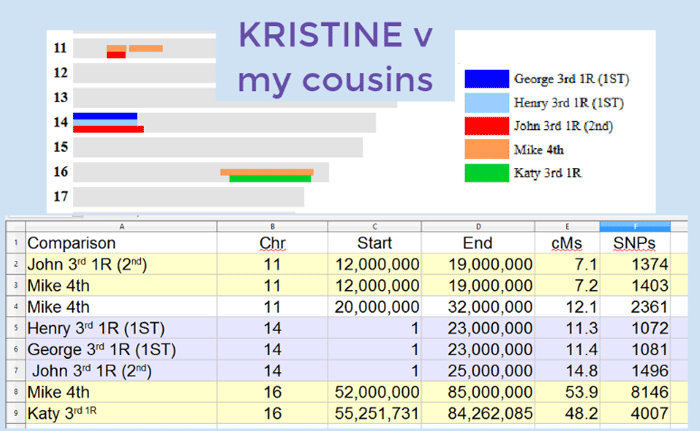
I also convinced another Wold cousin to test in order to help out. This was a good thing since Kristine shared no DNA with my nuclear family but she did share DNA with my first cousins (brothers Henry and George), my 2nd cousin (John), and much DNA with the tested Wold cousin Mike (my third once removed).
Every place you see two parallel colored lines in the diagram above, two of my Wold side cousins are matching Kristine’s DNA and each other in the same spot. That is triangulation. Because her matching DNA triangulated with several different cousins in three spots, we are now confident that Kristine shares our gg-grandparents Anna and Jørgen Oleson Wold (click here for the full story). [addendum 19 Oct 2015: this diagram was updated to the one used in my triangulation presentation which includes cousin, Katy, found via DNA testing after this article was originally posted. She is a 2nd cousin 1R to Michael. The details of this triangulation are shown in my triangulation presentation, slide 8 on]
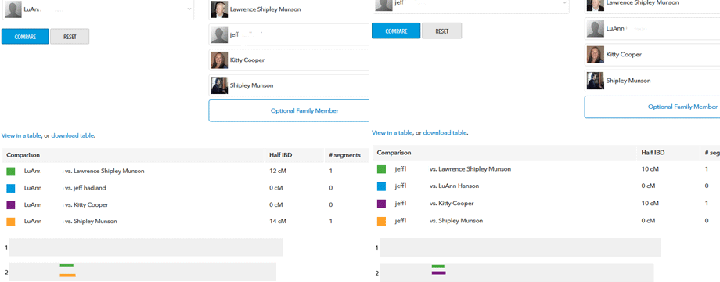
Looking through the master spreadsheet for my Dad, I see a reverse case. Jeff matches my Dad and me from 46M to 57M on chromosome 2 for about 10cM while LuAnn matches my Dad and my brother from 46M to 58M also on chromosome 2. Even though they each match Dad in the same spot, they do not match each other there. Plus one matches me and not my brother while the other matches my brother and not me. This is a good example of a match that is proven to be from different sides, one maternal and one paternal, and thus different ancestors. In other words, a pair of matches at the same spot that did not triangulate with each other, but did triangulate with other family members.
So how do you do a triangulation?
The easiest place to do a triangulation is GEDmatch.com – there you can use the one-to-one comparison tool to compare A to B, B to you, and A to you and then see if all three share the same segment(s).
On 23andme it is also easy, provided you are sharing with both A and B. You use the Family Inheritance Advanced tool (under Ancestry Tools on Your Results) to compare all three. When you see that A and B match you, then you switch the person on the left to A or B and put yourself on the right with the other one. Now you can see if A and B match each other at that same spot. Below is the example with Jeff and LuAnn from the case previously mentioned. In the first one LuAnn is compared to my family and Jeff. Then I switched her and Jeff for the next compare.
UPDATE: 21-AUG-2020: You cannot do DNA triangulation at Ancestry because there is no display of the actual segments, known as a chromosome browser. However MyHeritage has automated triangulation included in its chromosome browser see https://blog.kittycooper.com/2018/04/myheritage-dna-matching-excellent-enhancements/
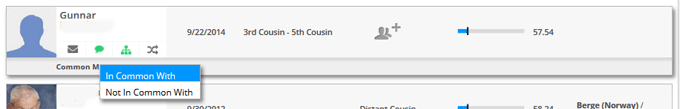
On Family Tree DNA it is more complicated to triangulate since you cannot directly compare A to B, you can only compare them to yourself or any other kits you manage. One possibility is that when both A and B match you on a specific segment you could contact each of them and ask them to check if they match the other at that location. A workaround that many use, however, is the In Common With (ICW) function. As in the example below, you can click below a person listed in your family finder match list and request to see all the people the two of you match in common by clicking Common Matches and then In Common With.
If A and B are in common with each other then it is likely that the segment where they both match you is a match. But it is no guarantee. The number I have heard is 90% but that is just an estimate based on experience; others say 99%. Below is a case where both Gunnar and Stephen match my Dad and they are each on each other’s in common with list. So a likely match.
Is there anything better? Well you can use other family members to check the match.
Since both my brother and I have tests at Family Tree DNA as well, I used the surname search to find these kits in my brother’s match list, then viewed them in the chromosome browser. If these two guys are a match, my brother will either match them both or none. Why you ask? Most likely, my brother only has either Dad’s paternal segment or maternal segment at this spot. DNA tends to travel in chunks and although it may get smaller as it gets passed down, it takes many generations for it to get more mixed up. Here is my brother’s view of these two in the chromosome browser:
By the way, neither of these fellows matched me. Have you noticed that this is the same spot on chromosome 2 that we were looking at for LuAnn and Jeff? So what else might we learn from this?
Well remember that I matched Jeff and my brother matched LuAnn so we each got different segments from our Dad here. Thus LuAnn must match Gunnar and Stephen as well. I am hoping Stephen and I will find this common ancestor for our 16.9 cM match, but no luck yet, and I have not heard back from the other two, a common problem …
Further reading on triangulation:
This blog post of Roberta Estes:
http://dna-explained.com/2013/06/21/triangulation-for-autosomal-dna/
This article at the ISOGG wiki:


This is very helpful. Is there a way to use the triangulation tool on GEDmatch rather than the one to one tool to compare results of five or six people? I have tried sorting out my results using FTDNA, DNAgedcom’s ADSA, and GEDmatch, and I always just end up confused and frustrated. I am just trying to compare results of my mother, brother, a known second cousin, and three or four close (2-4th cousins) matches about whom I do not know the connection. I just keep getting lost!
I did a post with details of the GEDmatch triangulation tool last fall http://blog.kittycooper.com/2014/10/new-utilities-at-gedmatch-tier-1-for-paid-members/
When looking at results, I find it easiest to view it from one person at a time. I keep a spreadsheet for each of my family members and it has a column for who else matches that segment plus I use a light background color to show Matching segments. Have a look a some of the posts here on using speadsheets http://blog.kittycooper.com/tag/dna-spreadsheet/
Or as Sara suggests, try genome mate
I also wrote a tool called Kworks which aggregates your ftDNA results with a matrix of the ICW. It works in conjunction with the tools at DNAgedcom to download a master ICW list and a master segment match list, see http://dnaadoption.com/uploads/DNAadoption/DNAadoption_files/DNAadoption/Jworks_Kworks.pdf
There is also a matrix tool at FTDNA that can be used to compare multiple matches to each other. The only thing here is that you can’t control the threshold for cm. It’s just a yes or no colored box in the matrix but still helpful nonetheless.
Another useful tool is Genome Mate.
The matrix tool is a really nice way to display the ICW information, Roberta Estes wrote a good post explaining it: http://dna-explained.com/2013/12/10/family-tree-dnas-family-finder-match-matrix-released/
The best I have found for this is a “Tier 1” tool (make a $10 donation to their efforts for a month of access) on gedmatch. Enter any gedmatch number and you will find a listing with each line containing 2 people that triangulate (each kit#, name & email), and the common segment for the 3 of you (chr, start, end & length).
Here’s my situation:
Family tradition says that Diana, Barbara, and Rose share a common ancestor.
Diana & Barbara match on chromosomes 1,4,6,9,10, 13-17, 20, & 22.
Diana & Rose match on chromosome 9
Barbara & Rose match on chromosomes 3, 4, 7, 10, & 16.
So, Rose is related to both Diana and Barbara, but since no chromosomes are shared in common among all three, does that mean Rose is related to Diana via a different ancestor than she shares with Barbara?
Triangulation proves a common ancestor but the lack of triangulation does not prove that there is a different ancestor. When LuAnn and Jeff did not triangulate in my post, there was triangulation with other members of my family to get to the conclusion of different ancestors.
So your case does not prove that Rose is related via a different ancestor. You need more data (more cousin tests) to prove or disprove that theory.
Well, I do have two more cousins involved.
Diana & Sylvia match on chromosomes 4, 6, & 9.
Barbara & Sylvia match on chromosomes 2, 6, & 20.
Rose & Sylvia do not match.
Doug & Diana do not match.
Doug & Barbara match on chromosomes 1, 6, 20, & 21.
Doug & Sylvia match on chromosomes 1-3, 5-6, 11, 16-18, & 20.
Doug & Rose match on chromosome 13.
One potential problem is that Doug, Diana, Barbara, & Sylvia’s trees have more than one ancestor in common.
I am feeling pretty lost and don’t know what to make of all of this. Thanks for your input.
Ciao sono Rosaria su mhyeritage ho molte corrispondenze e anche triangolazione con molte persone che anche se vivono in tutto il mondo hanno origini albanesi.i segmenti quelli che triangolati ha volte e 9cM e poi cambia quando aggiungo altre persone.che vuol dire?io sono del Sud Italia che è moto vicino ha paesi dei Balcani e su mhyeritage ho il greco albanese grazie.
Hi Rosario,
The triangulation changes when you add more people because not everyone has the exact same piece of triangulating DNA. Some may have more and others less.
As to the ethnicity, that is not an exact science and MyHeritage is the least accurate with it. Albania is just across the water from Italy so there are likely many common ancestors a long long time ages.
I don’t mean to presume on your good will to assist me, but if you have time to check out my following posts on the ISOGG Facebook page concerning this group of cousins, I would appreciate your thoughts:
https://www.facebook.com/groups/isogg/10153086774407922/
https://scontent-atl.xx.fbcdn.net/hphotos-xpa1/v/t1.0-9/10462516_10204808771113580_5787956519899259404_n.jpg?oh=a37b3822151266e1124fce93355f21ef&oe=5594C094
Terry I took a quick look and nothing jumps out at me. Get more people tested. I will comment over at facebook.
And I do apologize, I cannot afford to spend much time on individual cases that are not my relatives or clients. But I do enjoy helping as I learn from every experience.
Ciao Kitty,sono marius ho una triangolazione DNA con me e tra loro sul cromosoma 5,loro hanno origini nordafricano ma vivono in altri nazioni.il segmento DNA triangolato e di soli 8cM.questo gruppi e di 6 corrispondenze che vuol dire?
Marius – That is a very small segment but if it triangulates it is most likely from a distant ancestor to all of you. I have found with such small sized segments that the common ancestor can be as far back as the 1400s or further. Or as recent as the 1800s. No way to know without detailed family trees.
Gilda ho 15corrispondenze dna con triangolazione con almeno 14 tra loro e me alcuni tra loro sono mezzi cugini primo grado tra loro tra loro sono anche secondi 3 4 5 cugini.con me sono tra4 5 cugini.Tu pensi discendono da uno ho più figli degli antenati comuni?
Having more than one common ancestor makes it more difficult and more complicated. The only time you can be confident there is a common DNA ancestor is when 3 people match in the same spot.
Do Doug, Sylvia and Barbara all match in the same place on chromosome 6? Do Diana, Sylvia, and Barbara match on the same spot on chr 6? If not then you cannot conclude anything from what you are seeing yet. Get more cousins tested. Getting the oldest generation to test can be very useful.
What is your objective with this testing?
Pingback: Noli Irritare Leones » Some links that I have already posted on Facebook, and some that I haven’t
Pingback: 2nd Great-Grandfather William Alexander McNamara, First DNA-Identified McNamara Ancestor
I have read your blog on autosomal dna and am still having difficulty identifying ancestors. I have tested5 family members and have 2 additional kits for my sister and my aunt. I have to say my dna is confusing particularly since an ancestor in my mother’s line appears to have married an ancestor in my father’s line. I thought this was unlikely since my mother’s line which matches appears to be irish and my father’s line is strictly polish. I have found however a story by several of my dna matches where a polish ancestor of their’s fled Poland by swimming a river and then married an irish girl. It seems likely by triangulation that the irish girl is a descendant of one of my irish lines however it has been extremely difficult to link lines with irish records. I am in general having difficult identifying chromosome levels with ancestors as many of my links where I know a MRCA are back in generation 7 or beyond with my cM match <5 although we may match on 5 or 6 different chromosome levels Is it possible to identify chromosome matches at a smaller level mostly if you also have a larger match with another individual. If you have identified an individual through a larger match, does it then stand to reason that the smaller match must also be that surname. I have started using the X-chromosome more and hope that I can identify individuals easier. Helen
I have a basic concept question about autosomal testing and chromosome segment matching. Can the segments of each chromosome be mapped as unique to either the maternal or paternal side, or are all these segments mixed up and you need to still determine which side someone comes from by comparing trees and triangulating with ancestors from each side? An example, when I look at myself, my mother and my aunt (my aunt is my mother’s half sister-they share the maternal line). The segments we all share in common on each chromosome must come from my maternal grandmother’s ancestors- the line we all share. If I look at any others who share parts of the segments we three share- then can I assume they are through my grandmother’s ancestors? 11% of my DNA matches my aunt, so that 11% we share through my grandmother’s ancestors. Does this mean that anyone else who matches us in those segments must also come from my grandmother’s side? An answer to this would help me better understand the whole idea.
You need to always use your matches to others to map out your chromosome. As I say in the article above “the testing mechanism cannot differentiate between the two” chromosomes, paternal and maternal.
Having a parent tested does let you see which side a match is from. And a half aunt is very helpful. Yes, your common matches with her must come from your maternal grandmother’s ancestors unless it is a false match for them which can happen with smaller segments. See my post at http://blog.kittycooper.com/2014/10/when-is-a-dna-segment-match-a-real-match-ibd-or-ibs-or-ibc/ for an explanation of false matches
Man.Ciao ho notato su mhyeritage che ho corrispondenze dna da germania ma poi vedo il loro alberi ma hanno origini dai paesi est europa e ho anche altre corrispondenze dna che anche da norvegia ho olanda ecc ma hanno sempre origini del est europeo.le corrispondenze non sono grande dna totale ma due fratelli il fratello mi corrisponde con dna totale 19cM con 3 segmenti tutti tra 6cM mentre sua sorella ha più di 20cM totale dna con 4 segmenti dna naturalmente con triangolazione dna lui e la sorella.poi un altra corrispondenza sempre da germania ma sempre con origine est europa dna totale 23,3cM con 3 segmenti dna con il segmento piu grande di 9,4cM con triangolazione con altre corrispondenze.
Well there has been much migration in recent and less recent times from Eastern Europe into Germany.
Through 23andme I learned I carry the Cystic Fibrosis CFTR gene mutation F508. This is on Chromosome 7 in the 117,xxx,xxx region. With ADSA I found I have a triangulated group in this region. My question is this: in the absence of informative ancestral trees, would the chance that any one member of the triangulated group also carries the mutation be 50%? Since there is no way of knowing if I or any of them have it on the maternal or paternal chromosome, right?
Melinda,
You are correct, there is no way to tell from just your test and I presume neither of your parents is tested to isolate the side that way.
But if you have other cousins tested who match you on that segment then you may be able to infer which side the CFTR is on from the cousin’s test. Then if you can compare your matches to your cousin (or have her send a screenshot of her ADSA results at that position) you can tell which ones may have it as well
Also my parents are deceased and untested.
I got a question. I have a relative on gedmatch who apparently shares enough autosomal DNA to be considered not just a cousin but a full sibling. Unless my parents adopted out some unknown sibling of mine, I possess one full sibling & one female half sibling [the relative is male apparently]. 3523 versus the 2678 I get with my sibling, 1.0 generations versus 1.2 generations.
Thing is, is this relative doesn’t share any chromosomes with myself or my sibling. Nor for that matter grabbing a bunch of random relatives and using gedmatch’s chromosome segment matrix anyone else.
So is he a relative or some glitch?
Sounds like a glitch. Contact GEDmatch at gmail dot com with the kit numbers
Perry.su mhyeritage avevo una corrispondenza e insieme avevamo altre corrispondenze in comune adesso su mhy questa corrispondenza non si vede più nella lista delle mie corrispondenze e di altri.forse ha cancellato il suo kit?grazie.
Yes, sadly he probably deletes his kit. You could ask MyHeritage support yo br sure.
Albert.su mhyeritage ho 4 corrispondenze dna e Triangolazione tra di loro e me il segmento piu grande che triangolati è di soli 7,1cM tra loro qualcuno sono cugini più vicini tra di loro mentre con me mhyeritage dice che sono cugini tra 4 ho 5 grado.ma fino ha ora non ho trovato nessun collegamento.forse e piu indietro?loro vivono in stati uniti ma hanno origini ungheresi io sono italiano.
Yes a segment that small can be much further back and hard to find. You might try uploading to GEDmatch and checking that segment there (requires a tier 1 membership)
Kitty, Thank you very much for a very helpful post regarding triangulation. It was referred to me on Genetic Genealogy at Facebook.
In your piece, there is a dead link to your “triangulation presentation”. Are you able to forward a good link to that, please?
Thank you! Rick
and thank YOU for pointing this out
All my presentation slides are always at http://slides.com/kittycooper/ but I did just update the article.
Kitty, I am trying to figure out something on the Triangulation tool. I have a brother and we share the same parents. I also have a half-sister, and we all three share the same father. Our father has passed away and I would like to try to map his DNA. Could I use the areas in the Triangulation results and where we all three match, would that be on our father’s side? Or, is there more to it than that?
I love your new DNA MAP tool on Genesis!
Thank you!
Andria
Andria –
Anywhere your half sibling matches either of you is from your Dad, not just the triangulated segments.
You can create a pseudo kit for your Dad using the Lazarus tool which is Tier 1 and I hear that another tool to recreate him may be coming out any day now …
In general, how far back can one triangulate? I am trying to triangulate back to 4th and 5th great grandparents, without much success.
The further back, the harder it is. You need lots of descendants to see any triangulations. Think about how little DNA 4th and 5th cousins share and realize that you need to be lucky to get descendants inheriting the same segments.
Kitty, Can i share DNA with several matches on the same segment and not have triangulation.
Phil, you can share DNA with 2 matches where one matches the segment from your Dad and the other from your Mom so they do not triangulate. A third person would have to triangulate with one of them unless it was a false match (usually smallish) see http://blog.kittycooper.com/2014/10/when-is-a-dna-segment-match-a-real-match-ibd-or-ibs-or-ibc/
When looking at a possible triangulation on FTDNA, where one of the overlapping segments goes from, say (using simplified starting and ending numbers), 1 to 10 and the other goes from 2 to 11, how can you tell the number of overlapping cM? (MyHeritage shows the number, but I can’t see how to figure it out with FTDNA.)
In the case you describe the possible overlapping segment is from 2 to 10, however there is no one to one correlation between the SNP count and the cM. Read this on that topic:
https://isogg.org/wiki/CentiMorgan
Plus the problem is that you cannot tell whether it triangulates at family tree DNA. You need to both upload to GEDmatch or MyHeritage to figure that out.
I need to prove DNA to my father who has been deceased since 1988. He is not on my birth certificate but he and my mom told me that he is my biological father and I had a relationship with him while he was alive when I turned 19. I did my DNA on Ancestry and my DNA matched my father’s brother and sister’s children. I also match my grandmother’s brother’s granddaughter. I am told that Ancestry DNA can be misleading and wrong. How do I further prove that I am a child of my father through his brother and sisters children?
Whoever told you Ancestry can be misleading was incorrect. They probably were thinking of the ethnicity estimates not the relative matching. The amount of DNA you share with relatives is very accurate, however the interpretation of that number to an actual relationship can be fuzzy since many relationships share similar amounts.
In your case, with so many paternal relatives tested, you (or a genetic genealogist) can likely build a model using WATO (what are the odds?) to prove that he is your Dad. I will send you a private email as well.
Hello, I did hire a genetic genealogist and she did build the model using ‘WATO. My biological father adopted a daughter (she passed) and she worked for Planned Parenthood her whole life and did not have a husband or children. The only heirs would be my biological father’s brother’s and sister’s children and the are all about 80. I did a hearing for the estate in Manhattan, NY and they are continuing the hearing and I need to provide more information as they do not want to use Ancestry DNA’s matching and said I would need to have a blood test to prove DNA. I don’t have anyone who would be willing to do DNA from my father’s family. My daughter and my grandson match the DNA matches with the same family members from my dad’s side. I think one of the most telling proofs is my DNA match to my dad’s mother’s brother’s granddaughter. She is a match with me and my daughter and grandson. My older son also has matches with these people.
Sounds like you need to talk to a lawyer who is familiar with DNA testing. Any chance your Dad could be exhumed for testing?
Ciao.sono Lara.su mhyeritage ho molte corrispondenze con un gruppo di 8 matches con triangolazione tra me e tra loro .il segmento piu grande che strangola con loro e soli 7,5cM.loro su mhyeritage e ho contattato una corrispondenza e mi ha detto che i suoi genitori sono dei nomadi.che significa?ce poi sempre di questo gruppo altre persone delle stesse gruppi genetici di queste popolazioni.
Lara,
Sorry I do nor know what he means by nomads, you would have to ask him.
Lara.mi spiego meglio,io su mhyeritage ho un gruppo di 16matches e ho delle triangolazione sul cromosoma 16,tra me e loro si formano parecchi gruppi tutti sul che 16.il segmento triangolazione ha volte e 8,4cM cambia quando inserisco più matches nel browser .il fatto dei nomadi queste persone hanno nelle loro stime etniche hanno collegamenti con popolazioni dei rom est europa.ho contattato una matches e mi ha detto che i suoi genitori hanno questi origini da queste popolazioni.anche altri dai loro cognomi hanno collegamenti sempre con queste popolazione.sono cugini più vicini e più distante tra loro e con me secondo mhyeritage sarebbero tra il 4 5 grado.in questi giorni ho avuto un altra corrispondenza di soli 8,4cM che è sempre collegata e triangolazione con questo gruppo che già ho triangolati.e questa nuova corrispondenza e sempre est europa ma vivono in tutta Europa.
Andreis leggo molto i tuoi post.grazie.su mhyeritage ho 6 corrispondenze triangolazione tra loro e me il segmento Tr e di soli 6,6cM ma ha molti più di 3000più di sono.loro vivono in Norvegia ma hanno nei Lori alberi antenati da ex Jugoslavia.può essere anche se e piccolo il segmento una parentela?
Sheila.ho una triangolazione con 11matches su mhyeritage il segmento triangolazione e 8,7cM ma non riesco ha capire come posso essere imparentati.forse ce qualche linea nel mio albero che non so?
That small a segment could be very far back or … look at those 11 matches and see if you can figure out how they are related. Then maybe look at that line for an ancestor of yours or perhaps a neighbor…
Andreis volevo dire che ho una triangolazione con il segmento triangolati di soli 6,6cM ma con più di 3000 dna.scusa prima ho scritto male.
Dalila,ciao kitty ho contattato il supporto mhyeritage riguarda corrispondenze di cM piccoli che si triangolati tra me e loro e mi hanno detto che devo stare attento con le pi se perche potrebbero essere casuali ho ibs ho dicono di esaminare bene i documenti e altro ecc.
Good advice. Small segments can be false or too far in the past to find.
Brenda.che vuol dire ibs segmenti dna grazie.
Brenda.che vuol dire segmenti ibs che però triangolati tra me e tra loro grazie.
Robert.Ho su mhy 2 corrispondenze che abbiamo in comune altri 14 corrispondenze e triangolino sul che 17 però 2 di queste corrispondenze hanno anche segmenti sovrapposti su un altro cromosoma il 4 ma mhyeritage non segnala la triangolazione uno ha il segmento 7,4cM un altro ha il segmento 6,7cM ma con 3000 sono e strano sul cromosoma 16 queste 2 corrispondenze triangolati con altri ma loro due sul cromosoma 4 non lo fanno.