Much of the work to build tools and write articles to help testers with their DNA results has been done by citizen scientists, bloggers, computer programmers, and scientists from other fields like Andrew Millard (behind the WATO math). In an exciting development, Amy Williams, a computational biologist at Cornell University, has built a few DNA tools, with more to come, and started a blog at https://hapi-dna.org/blog/
Her blog article titled “How often do two relatives share DNA” is particularly interesting. It includes the beautiful chart shown below which is created from simulations. Click it to go to the actual page where you can mouse over the columns to get the detailed numeric breakdowns.
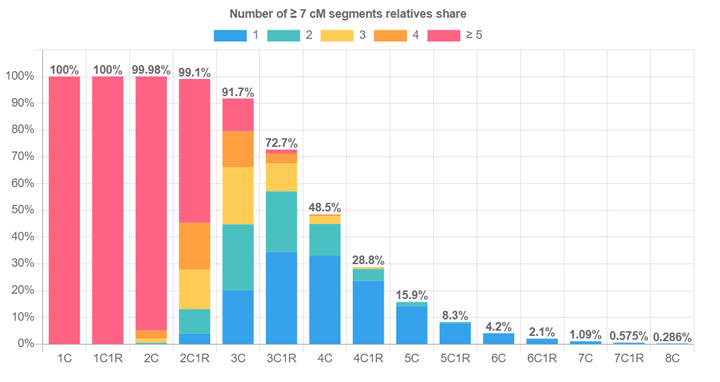
Chart of How Often 2 Relatives Share DNA from https://hapi-dna.org/2020/11/how-often-do-two-relatives-share-dna-2/
The other article on her blog has a detailed explanation of what a centiMorgan is, the measurement used for DNA segment sizes (click here). I usually recommend not worrying about the exact definition since it is a measure of the frequency of recombination rather than a physical length. It is important just to know that there is not a one-to-one relationship between the cM and the sizes shown in the chromosome browsers. On the those charts, the same cM amount looks smaller at the ends of chromosomes than it does in the middle because recombination is more active on the ends.
The final statement of that article is: “In an upcoming post, we’ll talk more about cM lengths of DNA and how recombination leads more distant relatives to share fewer segments that are also on average smaller than those that close relatives share.” Something to look forward to!
Now to take a look at the tools that are available there so far.
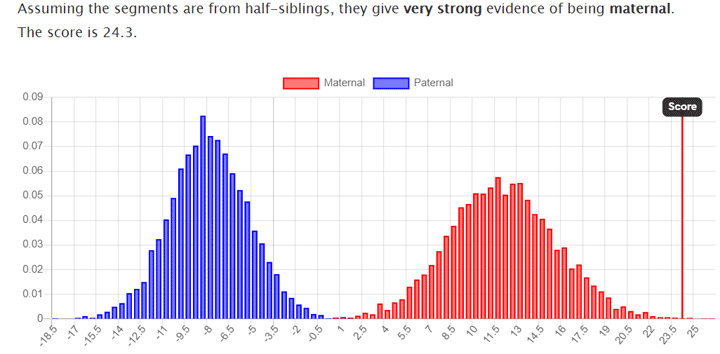
Of particular interest to adoptees is the maternal versus paternal predictor for half siblings or grandparents. I tried it out on a number of half sibling pairs who I have helped in the past.
Here is the prediction created for a brother and sister who share the same father but have different mothers, using the comparison of their segment data from 23andme:
However I discovered number of minor usage issues when trying to use data from the different DNA testing sites.
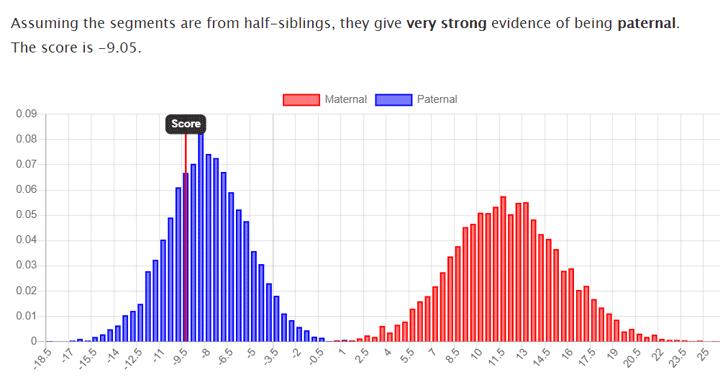
You can cut and paste from the GEDmatch one to one comparison, but if the kits are migrated kits, that is kits from the previous version of GEDmatch massaged into the new template, the result will not be accurate. Best to reupload the DNA data. This chart is the same brother and sister who share a father as the above, but this time from a GEDmatch one to one using their migrated kits.
You cannot just cut and paste the segment data from the page at 23andme or Family Tree DNA.
- For 23andme you have to copy the data to a spreadsheet, then sort it by chromosome and start point and then copy that data into the form. That is because when you have more than one segment on a chromosome they are not listed in order.
- For Family Tree DNA you have to download the data, then just copy that data from the spreadsheet since it is already sorted
- If you try to cut and paste from the chromosome browser comparison at MyHeritage the spacing is not preserved so it fails. You have to use the little advanced options button below the image on the right to download the segment data then just copy that data from the spreadsheet, it is already sorted.
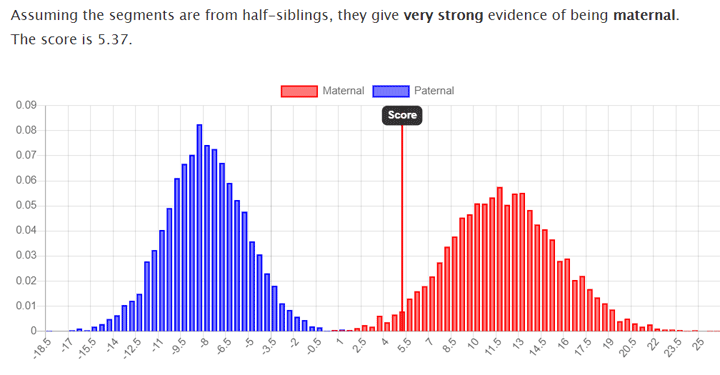
Known half siblings with the same mother from GEDmatch new template, low amount of shared DNA 1368.8cM
This answers a question I have had for awhile. I have always wondered if having one segment was enough to consider realistically a match. I have an unknown gr gr grandfather who I have matched to a line of DNA cousins. This helps me feel like it is absolutely the right person. This is a wonderful tool.
You might enjoy my search for my gggrandad’s daddy here.
https://blog.kittycooper.com/2020/05/who-is-my-great-great-grandfathers-daddy-a-thrulines-experiment/
Yes that far back single segments are the norm.
Kitty, thanks for sharing that chart.
That 2 relatives share DNA chart is the best representation I have seen of stats that have been around for about a decade or more. More great stuff at https://isogg.org/wiki/Portal:Autosomal_DNA under internal links to pages on Autosomal stats, Cousin stats and Identical by descent.
Working out whether you might share >some< matches with DNA relatives also requires input from how many relatives one typically has at each level: C, 2C..5C and so on. And then merging those two together.
From memory, someone said the sweet spot was around 5C. The chart above shows we only have a 16% chance to match to any specific 5C directly. But we might connect via an intermediary.
Maybe Amy Williams can help us to see that better.
Another good set of simulations from Amy Williams showing the likelyhood of sharing DNA with half relatives https://hapi-dna.org/2020/11/how-often-do-two-half-relatives-share-dna/
My apologies, need more instructions. How is the score determined? How does one cut and paste segment data to this chart? It’s too difficult except for computer scientists. Would like to use it. Thank you for this.
Shelley –
I don’t worry about the score myself, I just look at where the line falls in the resulting diagram but ask that question on Amy’s blog.
As to the cut and paste, you can only use it with the segment data which is not available on Ancestry. On GEDmatch do a one to one comparison using “Position Only.” Then highlight the data from the first segment listed through the last with your cursor, then control C to copy it, move to the blank form in that tool and type control V to paste it.
For how to cut and paste from a web page see https://www.computerhope.com/issues/ch001328.htm
The problems happen when copying from other sites because the segment data does not all stay on one line. Then you need to either edit it manually back to one line or use a spreadsheet program. Here is my post on spreadsheets:
https://blog.kittycooper.com/2016/12/using-spreadsheets/