The same question seems to come up over and over again among those new to autosomal DNA testing. If I match A and B on the same segment why is that not enough to prove they match each other and we have a common ancestor?
The reason the ancestor is not proven is that you have two strands of DNA on each chromosome (remember there are 23 pairs of chromosomes) and the testing mechanism cannot differentiate between the two of them. So A could match the piece from your mother and B could match the piece from your father or one of them could even be a false match to a mix of alleles from both parents (see my post on IBC for more on that concept)
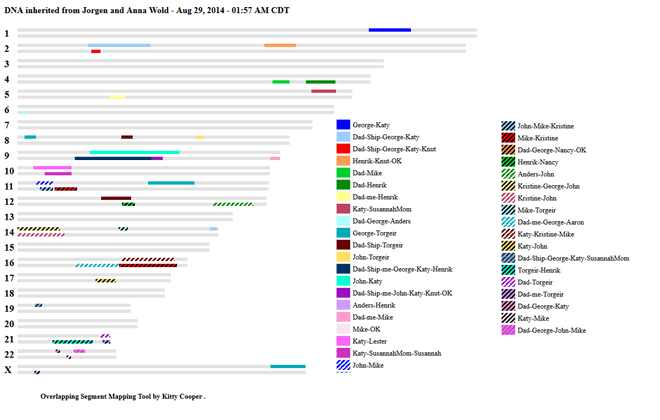
The way to prove the common ancestor is to see if A and B match each other in the same place that they match you. This is what we call triangulation.
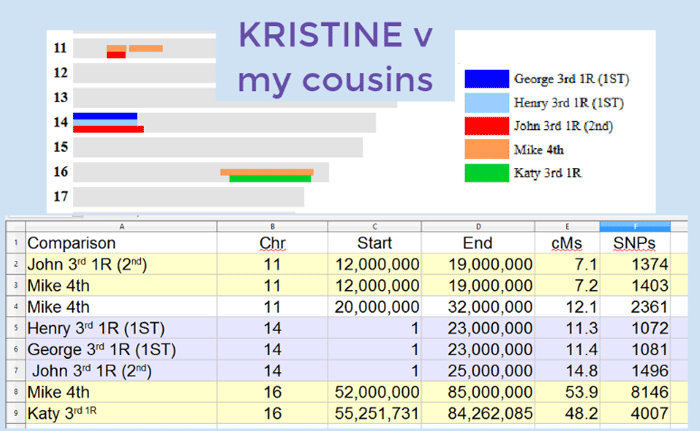
About a year ago I blogged about how, after many years, a change in spelling on the paper trail had led fellow genealogist Dennis to think his wife Kristine was perhaps descended from my great-grandmother’s brother Carl. To prove this I suggested he test her autosomal DNA.
Continue reading