UPDATE 14-AUG-2018: This tool has been superceeded by a far better one which will save the profile as well over at DNApainter.com
This tool is for the advanced user who has assigned DNA segments to specific ancestors. To compare segment data for multiple (possible) relatives use one of my other tools.
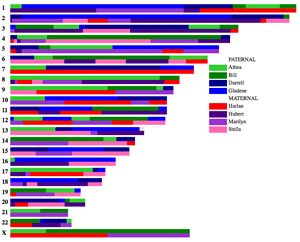 This program will make a pretty chromosome map from a CSV file which lists ancestral DNA segments from your known paternal and maternal ancestors, up to 20 ancestors on each side (you can have more if you specify your own colors).
This program will make a pretty chromosome map from a CSV file which lists ancestral DNA segments from your known paternal and maternal ancestors, up to 20 ancestors on each side (you can have more if you specify your own colors).
The secure URL of the tool is broken so use: http://kittymunson.com/dna/ChromosomeMapper.php
More about it with pictures in my August 3, 2013 blog post and more pictures, including the one to the left, in this September 2014 post.
Use my other tool, the segment mapper, to make a chart of overlapping segments from relatives or other DNA matches for up to 40 possible relatives (or more if you specify the colors). And use my one chromosome mapper for a detailed look at overlaps on a single chromosome.
The Chromosome Mapper requires the column headings and contents as listed below, case sensitive, in any order, in your CSV spreadsheet. It is intended as a tool for picturing your ancestral DNA as determined from known relatives as opposed to showing your DNA overlaps from unknown relatives.
Common Problems
The most common problems are not having the correct column headings (see below) . That includes using different capitalization from the listed names. For example, someone recently used Side instead of side and the program did not understand that (yes I will add this soon).
The next most common problem is not having a correctly formatted CSV file. Try using this online tool to fix your CSV – http://csvlint.io/ – or use google docs.
Another problem for those who have created their own CSV files manually is that when a segment begins at the exact same location as the previous one ended, so the numbers are the same, the second segment will not be shown. Add 1 (or .1 if in millions) to the second number. Yes I think this is a bug that I should fix!
Mac users creating CSV files from Numbers or other programs have line endings that my program does not see correctly. To avoid that please read the article on creating a CSV for some work arounds.
Making an Ancestor Spreadsheet
To better understand the chromosome mapping concept, read the ISOGG wiki article on mapping. Also I have an article on making a spreadsheet of your chromosome matches on this blog.
Once you have a spreadsheet that you want to make a picture from, that sheet has to be saved as a CSV (comma separated values) text file in order for this tool to make a chromosome map from it.
Rebekah Canada did a really nice step by step tutorial of how to use this tool with screen shots on her blog, no longer available so click here for the archived version:
Current List of Required Columns in your CSV file for this Tool
| Column name | Alternate Name | Must Contain this Data |
|---|---|---|
| side | This column is optional, select which side (warm or cool colors) the segment appears on. To use it put the letter M or P or any words starting with those letters to indicate maternal or paternal side of your tree, this must be added by you to any CSV downloaded from 23andme or Family Tree DNA or DNAgedcom | |
| MRCA | Comparison or any column name you specify | Most Recent Common Ancestor(s) – this is the name to use in the chromosome map picture, you can tell the program to use a different column name for this. |
| chr | Chromosome | Chromosome number 1-22 or X |
| Start | Start point | The starting number for the segment location used by 23andme and Family Tree DNA, in the 23andme ancestry these numbers are divided by 100,000 so check the box for decimal if you are using that style of number. |
| End | End point | The ending number for the segment (see above comment). |
| cMs | centiMorgans (cMs) | The number of centimorgans in the segment, used for display purposes only. Since this is for display only you can put anything in this column for example I put the relative then a dash and the cMs |
| Color | color colors | This column is optional, you can let the tool pick the colors; but if you want different colors then put any color name from this list: http://www.html-color-names.com/color-chart.php in the color column the first time an ancestor appears in your CSV. The color name must be exact and case matters. Optionally, you can use a hex color like on the first page of this tool with a # in front of the hex digits. Warning, if you use one of the colors shown, it will also be used in order for another person, fix this by looking at the result and then specifying a different color for that person as well. |
Sample spreadsheet for this tool:
Programming is how I make my living so a small donation of $5 or $10 would be most appreciated, if you find this tool useful.
My paypal page is here: http://openskywebdesign.com/PaymentPage.html
73 Comments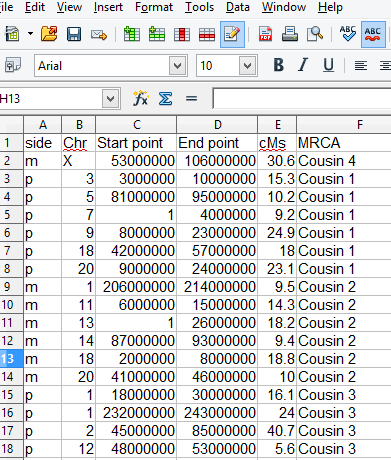
THANK YOU!
I have been thinking of this where to get a program, where to put the
matches together and here it is.
Have to try immediately next week.
I have a mac. I use Google Docs a lot. You can save spreadsheets created there as csv as well.
Kitty,
I just posted this on the Newbie list. Thought I should post here too.
Kitty kindly agreed to make this tool after I posted a request for a programmer to do just that. The idea behind it Is to be able to map KNOWN ancestor’s
segments. You cannot do this by simply downloading a .csv file and uploading to the Tool. And this isn’t for the casual DNA genealogist.
There are actually many ways the tool could be used as it allows you to label color segments and have them “plot” on a chromosome map.
1) So you could map by ancestry composition like the Ancestry Composition tool does at 23andme but you can include stop and start points which would be
helpful for comparing with matches.
2) You could paint “known” segments of your 16 gg-grandparents.
3) You can paint the MRCA segments for any match
Basically the tool gives you the power to create graphic representations easily via a .csv file.
Now the HARD PART. (And although I save match information getting this to work was not easy) First of all you must use a program that allows you to save a file in a .csv format which is basically comma delineated data. I could not find where to do that on my Open Office program (maybe EXCEL Office is easier to find) So I switched to using Mac program “Numbers” which allows you to save file in .csv. The suggestion above to Use Google docs is a great one.
Now what to save. Yes you can dowload data in this format. You can download all your match information from 23andme or 5 (10 I just learned) at a time in FTDNA or use DNAgedcom.com to download from both easily. BUT that gives you all your data and matches not the ones you need or have identified. The way I have tracked data from matches is each share at 23andme I compare with known relatives and plot that info in a spreadsheet. When I know the MCRA I note that or where I suspect the connection. At 23andme I look at their ancestry composition for the shared segment for clues. If the match and
I both share German-French on that segment that’s a clue. Over time you build up a list of identified segments and now you can use the tool to create a
Chromosome Map.
if you are just starting out you may not have ANY KNOWN segments thus you would have a blank Chromosome Map.
As to the nuts and bolts. I just copied the headings directly as Kitty has them and pasted them into a new spreadsheet in the first row. Then I copied and
pasted the data into the new spreadsheet from my tracking sheet. For some segments I did not have the cM length so I just faked a length (entered an
estimated value) which worked fine. Chosing colors was a bit tricky as you have to place your data in a way that selects the color you want. Perhpas in later
versions there will be an input for chosing colors.
For those of us wanting such a tool this is FANTASTIC so Thank you Kitty! But like all tools it takes some practice to be able to use it effectively and you can’t use a paint brush without paint. So if you want to use the tool you’ll have to start the painstakingly slow process of tracking matches and identifying segments. Which brings us round to the frustration of not having matching segment data at Ancestry where I have nearly 100 identified matches!
I hope this helps some of you understand the genesis of the tool and how to use it.
Kelly
Thank you so much Kelly for explaining this so well.
Awesome tool Kitty. Well done.
There is a bit of a problem for those who don’t know their parentage as it won’t run if you don’t declare a p or m in the side column. Perhaps the instructions should say “If in doubt default the lot to p”?.
Also an add on to your documentation for Mac users, when saving from Numbers it offers 3 format choices for the csv format It must be the windows format to make it work. Also the capitalisation and spelling on the headings must be as written exactly or it won’t play.
The next release will do away with the “side” requirement by just assigning alternate sides to your data and allow you to select the colors and so include more names that way. But read what Kelly wrote above for more about the original purpose of the tool.
I also have an article here about making spreadsheets http://blog.kittycooper.com/2012/11/making-a-spreadsheet-of-your-dna-matches/
Thanks for the tool.
My interest is taking a group of matches that I don’t know how they are related to me. So, I’d like to compare the chromosome matches one or more unknown matches to one or more known matches. By seeing where the unknowns are in common with the knowns, I’d infer who the common ancestor is.
Perhaps in a future version, any chromosome that has overlaps from different people, the graphic would just get wider to accommodate the overlap.
Help
I do not get any results. I have exported file from 23 and me. It is accepted and when it downloads to 100%….contacting Kitty Munson. I have tried this several times.
Thanks!
Please read Kelly’s comment above. Please check that your file is a CSV. You can do this by opening it in notepad or other text editor. If it is not plain English with many commas then it is not a CSV.
I will write some documentation on creating a CSV soon since so many of you have trouble with that. It is particularly tricky on a Mac.
Other users can create a CSV but then leave the side and MRCA columns blank and are surprised when the tool finds nothing to plot!
Would it be possible to add one more column to the CSV file that would permit adding a note to the MRCA pop-up? This would allow me to give all segments from a particular great-grandparent the same color across my map, but then add a brief comment like “COOK/BIRDSALL” to only one of those segments (to pick one of my ancestral couples, way back in the 1600s, where I have identified a common ancestor at 23andMe), without exhausting the color palette.
Good idea! I will add that to the enhancement list. The column will be called “note”
Elizabeth –
Because the number of centimorgans in the segment is used for display purposes only, you can put anything you want in that column. For example I put the relative’s name then a dash and the cMs. So try that for your comments.
Kitty,
Great job. I have a suggestion. I like the (M)aternal and (P)aternal side, but it would be awesome to add an (U)nknown option. This will make a third row on each Chro. This way the unknown is not covered by the M or P. Use a gray scale or paterns.
Good idea. I have also had a request to allow “Both” when an ancestor is from both sides so the third line would be “both” or “unknown.”
I also plan an additional feature in this tool to allow up to 5 or 6 tracks, no sides, to look at overlapping segments graphically. Probably release 3 or 4
Awesome Tool, Ms. Kitty!!! Thanks so much for creating it–donation on the way!
I’d really like to try this but am wondering since we are uploading our DNA to your program, does that mean that you have a copy of our DNA? I was hoping that I would be able to download the program to my computer and then upload my results. Is this possible?
Jan you are not uploading your DNA to this program. Rather you upload a CSV version of your spreadsheet of segment locations of DNA from specific ancestors. This file is not kept here after being used for the display.
The advantage of a web based program is that it works on Macs, PCs, tablets, …
I just released a new version of the mapper which will work without a side column by randomly assigning sides.
A suggested use is to take your FIA file from DNAgedcom and sort by largest segments and then run that file in this tool. You will see your top 20 matches.
Kitty,
First of all, thanks for this mapping tool. I feel it is going to be a valuable tool. I have two quick questions. After downloading the spreadsheet and getting the headings correct, I end up with each of my matches listed along with each chromosome they match segments on. When I find a documented connection to this person (let’s say it is Smith born 1830) do I add this MRCA to each of the chromosome matches? How do I determine which chromosome match is correct for Smith? My second question is, as I get additional matches on FTDNA FF, do I create a new spreadsheet (this overwrites all the headings etc I have prepared) or should I enter them manually?
Thanks
David
David –
Assigning a DNA segment to your paternal or maternal side is called phasing. There is an ISOGG wiki explanation here: http://www.isogg.org/wiki/Phasing
If you do not have a parent to test, then 1st and 2nd cousin tests can be extremely useful for phasing.
As to whether to add an MRCA that is up to you. This is just a tool for turning data into a pretty picture of chromosomes which can be used in many different ways.
Same answer for creating a new spreadsheet. If there is no additional data in your current spreadsheet then you can just paste in the new column headings on the new sheet. Column ordering does not matter to the tool. Personally I keep adding new data manually since I have much information in my current spreadsheets.
I am working on a new tool now, a segment mapper, which will show up to 60 people with matching segments on five lines within each chromosome.
For all of you who really wanted to use this tool to look at overlapping segments from unknown DNA relatives, I have now released a separate tool for that purpose! See http://blog.kittycooper.com/tools/segment-mapper
I recently did microarray to get transcript of Agave sisalana plant and full annotated it, now i am interested on chromosome mapping can you please help me ?
Having run the chromosome mapping tool, where is it to be found? If execution falls over for any reason, is there any error messages?
I have improved the error reporting for the segment mapper but possibly not this chromosome mapper. The results appear on your screen. If they do not there is an error in your CSV file. Please check that it is a CSV and that all the headings are correct.
I’m getting the error message, “Column heading MRCA is required.
Your file is probably not a CSV file please fix it and try again.” The file is a CSV–I tried saving it as such in both Excel and TextEdit–and I do have a column with the heading “MRCA.” I checked all of the column headings for accuracy and case. I’m using a Macintosh.
What’s wrong?
Excel on a MAC does not do the line endings properly. Read the article on making a CSV for suggestions on how to deal with this problem
http://blog.kittycooper.com/tools/making-a-csv-file/
Got it! Saving as “Windows Comma Separated (.csv)” solved the problem. Thank you!
P.S.: Here are the first two rows, the first version pasted from TextEdit and the second from Excel:
side,Chr,Start point,End point,cMs,MRCA
p,1,7414707,38986739,51.1,Baynes
side Chr Start point End point cMs MRCA
p 1 7414707 38986739 51.1 Baynes
Does anyone know if it is helpful to include the data of a sibling on this list?
Cara
You might try posting this question on the ISOGG DNA_NEWBIE list at yahoo. The idea of these tools is to use them as you please. Thus the answer depends on what you are trying to do. This chromosome mapper tool is for DNA segments you have assigned to specific ancestors but of course you can use it in other ways. I use the Segment Mapper tool with my brother’s data to look at known relatives, posted here.
My file used to work in the mapper tool. Updated it now it doesn’t. what could have happened? Please help.
Kathy, double check the column headings, doublle check that it is a CSV. If you are on a Mac the line endings can easily get mangled …
read http://blog.kittycooper.com/tools/making-a-csv-file/
I am offline or I would offer for you to send it to me. I will email you when I am online again if you have not resolved it
Having looked at Kathy’s file here is another tip for all of you:
When a CSV file does not work, try looking at it in a text editor like notepad to see what might be wrong.
In her case she would have seen lines with endless commas ,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,
So however she added those new lines did not work correctly, it added them all to one line perhaps
Hi I did dna testing to find my dad. I have all this testing and I do not understand how to narrow it down to find him and his family. Do you have any ways for me to narrow down all these people to help me get to the one person I’m really looking for. thanks for any help you can give.
Caitlyn it takes a lot of hard work to use this DNA testing for that purpose. There are many Search Angels who can help you with this. Please look at the resources I have listed for adoptees which can help you too http://blog.kittycooper.com/dna-testing/newbie-faq/#__RefHeading__792_704084449
Also read my page on DNA basics Which has a link to Kelly Wheaton’s excellent and extensive lessons on DNA
Is there any potential to use this if my test was done through AncestryDNA? They have no chromosome browser and this seems like a beautiful solution to my problem!
Another possibility if you have multiple kits at ancestry is to download the raw data for each and then use the tools at http://www.y-str.org/p/tools-utilities.html
There is a tool called autosomal segment analyzer there which you could use after using the convert tool on that site to convert your raw data to 23andme format
Yes but …. All my tools work with CSV files of segment start and stop points, not the raw data, so you would upload your raw data to GEDmatch.com once it is back to full service to get that information (and convince your closest matches to do the same)
Alternately you could transfer your ancestry results to family tree DNA for a small fee
I sweated over it and made a csv with 40 people, and assigned all my own colors, as I read it if I did, that I could have more than 20. But when I run it — it leaves out 20 people! What am I doing wrong??
I may have only added that to the segment mapper, I will double check the code and fix it today if so. Meanwhile I will send you an email so you can send me your CSV in order for me to use it to further test this, thanks!
all fixed now, thank you Linda for finding this issue
This is amazing!!!! I’ve been trying to do something like this by hand, but it was just getting too complicated. Thank you so, so, so much!!!
Thanks Kitty,
Worked like a charm. The error messages were most helpful. So far I have my mom’s DNA test and have phased my results at Gedmatch based on her’s. I did a very simple map showing my Gedmatch matches and whether they were on my mother’s or father’s side. It’s very helpful to see 945 lines of matches in one picture.
Next I’ll add my FTDNA maternal and paternal matches based on inferred connections between FTDNA and Gedmatch and remap.
Then I may map my wife’s DNA, but it would be based on her 1st cousin once removed as she hasn’t tested her parents. So her sides would be Lefevre/Pouliot vs Butler/Ellis. Her parents are Butler/Ellis and Lefevre/Pouliot are on the father’s side.
Very cool.
A classmate of my granddaughter has a Beauregard/Lefevre ancestry. I am interested in pursuing any connection between lefevre & pouliot that may shed some light on a possible conenction between that lefevre line and my granddaughter’s pouliot line. I understand that all pouliot’s are descended from Charles Pouliot of St Cosme de Vair. Is your lefevre/pouliot information available on line?
I’m building the spreadsheet required to use your Chromosome Mapper tool. Both my paternal and maternal lines are full of endogamy — how do I handle a quadruple cousin? Do I enter her data four times under the four different MRCA’s, or just once and leave the MRCA column blank, or what? So far I’ve identified 8 different cousins with MRCA’s (and no, I’m not AJ).
Thanks for any guidance/suggestions and for your wonderful blogs.
Julie
Julie –
My tools are for you to use as you wish. The intent of the chromosome mapper is to use to may ONLY those segments you are sure of. With endogamy it may take many cousin tests to figure out which ancestor gave which segment if it can be figured out at all!
I recently heard from a known 5th cousin that we share another set of ancestors so are 6th cousins as well! Now I am no longer sure which ancestor the second half of our large segment (17cM) match is from. The first half is shared with a cousin who does not share the other ancestors. I would guess the second half is from that other pair of ancestors but I will not know for sure until I find a cousin on that line only, who also has that match.
Hello Kitty
Thanks for the tool I have used it in the past with success but haven’t added ancestors for a while. I am using the same spreadsheets before (I think!) so not sure what is happening .
My kits don’t seems to be showing up on the right sides. I have M’s and P’s in the right places in the column ”side’. On my previous version I had headings of MATERNAL?PATERNAL. I don’t seem to have these headings now for either Maternal or paternal as they are shown in the example images.
Is it something about the order in which they appear in the spreadsheet?
Is this part of the changes? Is there something I can do to fix this?
Veronica
I’m sorry to sound clueless, but I guess I am, so…
I have downloaded my raw data from 23andMe. It gives only these columns: # rsid chromosome position genotype
How do I get data for the other columns you mention? Sorry if I’m missing something very obvious.
Thank you!
Prue
Prue,
These tools are not for working with your raw data, they are for displaying your segment matche lists. This particular tool is not for the beginner as it is for when you have assigned specific segments to specific ancestors.
The tool you want to be using is probably the segment mapper with the CSV with your match data not your raw data. You can get that from23andme in Family Inheritance advanced and from family tree DNA or GEdmatch via the tools at DNAgedcom.com (my next blog post soon …)
Kelly, how can I reach you to talk private?
Hi, I put the wrong name your name is Kitty?
I am having a small problem with my data. I have two csv files that are almost identical. One has an extra “bogus” line added. The one with the extra bogus line maps fine, but when I take out the bogus line, one of my entries for Chr 11 does not map. It is very strange.
I have checked my data and can’t seem to find an error. Any ideas?
I just started using the tool and am very pleased!
Thanks.
Leave the bogus line in 🙂 I will email you so you can send me the file …
Remember this tool is for when you have your chromosome mapped to specific ancestors. To look at overlapping segments from possible relatives use the segment mapper tool.
Kitty,
I am having the same problem as Veronica. Segments that I have identified as P are showing up on both the top & bottom half of the chromosome (and vice versa with M). So on the same chromosome, I have some of the paternal segments on the top half and some on the bottom half making it confusing to read. Any ideas on how I can change this so all my paternal segments are on one half and all my maternal segments are on the other half of the chromosome?
Kathy, Are you sure you are using the ancestor chromosome mapper? Usually when someone has this problem they are using the segment mapper tool.
It is the ancestor chromosome mapper.
Kathy,
Veronica was using the wrong tool and that was last July.
Since you are using the correct one please double check all your column headings. I sent you an email to send me the file.
It is very likely something in your data since no once else is having this problem.
And Kathy used the column heading Side instead of side so the tool did not think she had designated a side. I will add something under common problems.
And eventually add Side as an alternate heading to side
Hi Kitty,
Do you know why the 23cM segment appears longer than the 29cM segment on my chromosome 8?
Also, do you have any idea why my chromosome 19 appears to have a defect? (pun intended!)
http://users.ncable.net.au/~bwashing/BenWashington.htm
Regards,
Ben.
Ben
The mapper is mapping the base pairs which is a physical distance not the centimorgans which are a probability of recombination measure.
I will look at the chr 19 issue later
Hey there, I tested this out by creating a CSV file that includes a single DNA match but the resulting map is blank. I don’t have 23andMe and am confused by the MRCA column (why does your example show the cells with “cousin 4”, etc.?). I don’t have a CSV file to download with cousin matches but simply want to create a CSV from scratch as I come across/find matches to create a map of where my DNA on each chromosome came from. I have Ancestry DNA and use GEDMatch. So what do I put in the MRCA column? Can I just put the name of the ancestor? And why is my map coming up blank? Thank you!!
KH
It sounds to me like you want the segment mapper which is the one for visualizing your DNA matches
http://blog.kittycooper.com/tools/my-graphing-or-mapping-tools/segment-mapper/
This chromosome mapper tool is for when you know which ancestor gave you which bit of DNA and that is what the MRCA column is. You also need every column listed above (like side) with the exact spelling and upper/lowercase as listed.
I understand how all of this works except for one thing. Let’s say I have 5 people that match on the same segment and they all form a triangulation group. I can show on paper that the common ancestor couple is John Doe and Jane White. On your map you are mapping to specific ancestor. How do I know which one of the above couple the shared segment comes from?
You do not know which one of the couple that DNA is from so you show it as from the child you got it from or as from that couple.
You may prefer to use the segment mapper to show the segments from that couple.
Here is an example of a map I did of all the DNA from my WOLD 2nd grandparents:
http://blog.kittycooper.com/2014/09/segment-mapper-tool-improvements-another-wold-dna-map/
I’m trying to use the ancestry mapper tool to display my visual phasing grandparent’s results. I put the first 2 chromosomes in a csv file to test it out but I get a blank map. I used the csv lint and it checked out okay. I used apple numbers saved as a csv and also excel saved as csv, same results.
Please double check your column headings that they match the above. Computers care about upper and lower case and silly things like that.
I will email you so you can send me the file if you still need help.
The problem is the common one where files from a mac do not have the right line endings for this program. Try reading your spreadsheet into google docs and using that or …
See the suggestions in this article
http://blog.kittycooper.com/tools/making-a-matches-spreadsheet/making-a-csv-file/
Love it! Thank you so much for sharing.
I had started doing graphs using a “Numbers” spread sheet. I couldn’t figure out how to get the segments to start and stop, so I was adding each segment and the graph manually. Arg!
this is great i like it! Thank you so much for sharing.
I had started doing graphs using a “Numbers” spread sheet. I couldn’t figure out how to get the segments to start and stop, so I was adding each segment and the graph manually.
keep it up.
My great great grand mother was sent here from England because she was pregnant. My great grand father was born on a ship on his way to America. He was adopted. How can I find out his blood line? He still has 3 living grandchildren in their 90s.
John –
Do an autosomal DNA test for each grandchild or at least two of them at ancestry (probably on sale starting Monday). Take those DNA raw results and upload to family tree DNA which has a good selection of English testers. Also upload to GEDmatch and MyHeritage
Then look at the matches in common since those will be from the line of your great grandfather. The techniques used for adoptees are the same as any unknown parentage situation. See http://blog.kittycooper.com/dna-basics/help-for-adoptees/
for more help.
Hello, Kitty. This is an excellent tool. Thank you. I have made a DNA comparison chart similar to the comparison tool on Family Tree DNA using your chart. I would like to use the chart in a family history book I am writing. Please email me so that I can more fully explain. Thanks again.
I am using DNA painter and I hadn’t realised that you had been doing this for many years. What a wonderful technique – has helped me actually make useful use of my DNA data. Really loving becoming a chromosome painter. Thanks for all your work on this and giving it to the community. Really a great gift.