UPDATE 11-AUG-2018: ALERT!! a new improved version of this tool is now integrated with GEDmatch. No CSV files required; it can be run straight from the multi kit analysis (Tier 1) see https://blog.kittycooper.com/2018/08/new-compact-chromosome-browser-on-gedmatchs-genesis/
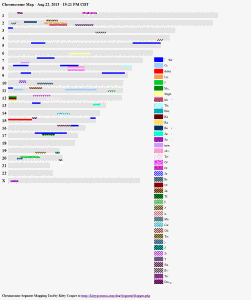
The purpose of this tool is to paint a picture of specific DNA segment overlaps from a CSV file of your segment matches. It does not work with your raw data but rather with your uploaded segment matches spreadsheet. That file is not kept on my site after the image is created.
The secure URL of the tool is not working so use this URL: http://kittymunson.com/dna/SegmentMapper.php
This page provides documentation on how to use the tool. It can use almost any downloaded CSV of segment data from the main testing companies as is or sorted.
Other Tools
For a detailed look at a single chromosome try my one chromosome mapper and if you want to paint a picture of which ancestors your DNA comes from, use my ancestor chromosome mapper.
Common Problems
The two most common problems are not having the correct column headings (see below) and trying to use this tool on your raw data (that will not work).
The next most common problem is not having a correctly formatted CSV file. Try using this online tool to fix your CSV – http://csvlint.io/ – or use google docs. Mac users getting errors please read the article on creating a CSV for some solutions.
Another problem is that a segment does not display. This is because it is on the same track as another person whose segment is a complete overlap to it. Assign a track number to one or both of them so they no longer hide each other (see the column documentation for track below).
How to Use This Tool
A suggested usage is to download your FIA file from DNAgedcom ; remove your immediate family, and then sort descending by the largest segments (genetic distance). It will also work with the Ancestry Composition relatives file from 23andme. If you keep a master spreadsheet of your matches from the different companies and GEDmatch, this tool can make a picture from that CSV as long as you use the correct column headings. Also if you want more than 40 people, you have to assign colors to the extra folk with the color column described below.
There are four tracks to display segments (but you can make it any number in the requesting form so 5 or 6 or …) on each chromosome which are assigned in the order that the people are listed. So the third person gets track 3, the fifth person goes back on track 1 unless you have specified more tracks. When overlapping segments end up on the same track the program shows what it can; you may prefer to use a track column to assign people to specific tracks so that overlapping segments end up on different tracks or just use more tracks. N.B. to see a specific chromosome in more detail, try the One Chromosome Mapper.
It was difficult to find more than 20 distinct colors that would work for even the color blind. So to go to 40 people I used a diagonal striped pattern when reusing the same 20 colors. Please let me know if any are difficult to tell apart.
Because the image created is html and images, you can cut and past it into an email but it is best saved via a screen shot (depress alt key and print screen) or save as a web page. The mouse-overs will not display unless you save it as a web page in which case you will also need the image files to see the stripes. Click here to download them in a zip file: stripeff99cc.zip. Put this zip file in the same folder as the saved webpage and then remove the stripeff99cc from the end of the folder name when extracting the images.
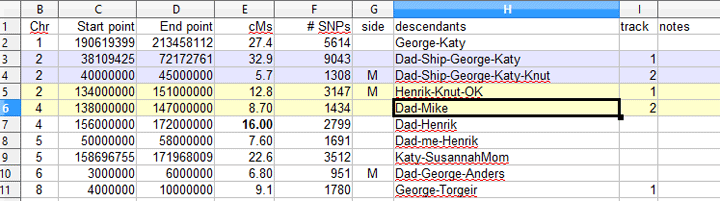
The Segment Mapper requires the column headings and contents as listed below, case sensitive, in any order, in your CSV spreadsheet
Current List of Required Columns in your CSV file for this Tool
| Column name | Alternate Name | Must Contain this Data |
|---|---|---|
| Comparison | MatchName or any column name you specify |
This is the name to use in the chromosome map picture, you can tell the program to use a different column name for this. |
| Chr | Chromosome | Chromosome number 1-22 or X |
| Start | Start point SegmentStartInMegaBasePairs |
The starting number for the segment location used by 23andme and Family Tree DNA, in the 23andme ancestry these numbers are divided by 100,000 so check the box for decimal if you are using that style of number. |
| End | End point SegmentEndInMegaBasePairs |
The ending number for the segment (see above comment). |
| cMs | Genetic Distance centiMorgans (cMs) SegmentLengthInCentiMorgans |
The number of centimorgans in the segment, used for display purposes only. Since this is for display only you can put anything in this column for example I sometimes put a note then a dash and the cMs |
| Color | color colors | This column is optional, you can let the tool pick the colors; but if you want different colors then put any color name from this list: http://www.html-color-names.com/color-chart.php in the color column the first time a match name appears in your CSV. The color name must be exact and case matters.Optionally, you can use a hex color like on the first page of this tool with a # in front of the hex digits. Warning, if you use one of the colors shown, it will also be used in order for another person, fix this by looking at the result and then specifying a different color for that person as well. |
| track | Track | This column is optional, you can let the tool pick the tracks which it will do in the order which individuals appear; but if you want to control which line of the graph a person appears on so as to eliminate an overlap that hides a result then you can use this column. See the example below which links to the article which discusses a “backwards” use of the segment mapper, that is mapping the descendants of an ancestral couple |
put image here
| Programming is how I make my living so a small donation of $5 or $10 via Paypal would be most appreciated, if you find this tool useful. |
42 Comments
I have a 23andMe FIA file from DNA tools (http://www.dnagedcom.com/), in .csv format. I get a message “Input file is not in comma separated format please save as a CSV file.”
Opening it in a text editor, I see it is separated by semicolons. Is there any advice as to how to convert the semicolons to commas? I do not have Excel on my Mac, but use Libre Office Calc.
Thank you.
Pinto I have added the semi-colon as an option for the CSV file. Try it now.
Hi Kitty, just got back to this. Thanks for adding the semicolon option. It doesn’t give me that error any more, but now says:
–> Column heading Chromosome is required.
–> Column heading Start is required.
–> Column heading End is required.
Please fix your CSV and try again
I’ve reworked the file so all information is in the 5 columns.
– Comparison column has Last Name, First Name of match, all in one column (previously in two).
– Columns are titled (case sensitive): Comparison, chr, Start, End, cMs (all aligned left, for good luck)
– I removed columns with my father’s name (the person being matched to), and a # SNPs column.
– I reduced the matches to 40 lines.
I still get the above error message. Any advice?
Thanks for any help.
chr needs to be Chr
Start and End should have been fine
it is not necessary to remove those other columns
Thank you for catching that. I changed it, still no joy. Changed to “Chromosome” and no dice with that either. Double-checked all spelling.
I will take a break on this for now, other duties are calling. Thank you for your help and providing this tool. I’ll try again another time.
I sent you an email requesting that you send me the CSV so I can see what is wrong. Are you on a Mac perchance? You can send it to me kitty at kittycooper.com
I really need help with this I am willing to pay for the help
It works as advertised. Being able to assign the tracks will be useful.
I downloaded my ftdna results from DNAgedcom. I sorted based on segment size to delete segments smaller than 3cM. I suspect the little ones are IBS and clutter up the display. I then did some gymnastics to sort the file according to greatest sum of shared DNA per person. I wanted to include the 8 relatives that I knew along with the 32 folks that had the greatest sum of shared DNA. So I copied to another sheet the entries that matched my goal, deleted the rows that I didn’t want, saved as a .csv file.
Thanks for your great tool!
Jim,
Even after deleting out segments smaller than “7 CMs” I still end up with over 500 matches. The largest being about 50 cMs. I would have to map 40 at the time. What I decided to do was delete all matches from the primary gedcomdna.com utility file greater than 7 cMs, then sort by Chromosome smallest to largest. I then cut and pasted the matches for each chromosome to a separate spreadsheet, sorted by cMs largest to smallest, added the appropriate column headings, and saved as a .csv file and ran Kitty’s mapping tool for each chromosome.
I stilled ended up with more than 40 matches on some chromosomes and the results end up a little cluttered.
Thanks Kitty for a great tool!
David
“TextWrangler” is an excellent and free text editor that is available in the app store.
The apple application “TextEdit” can also be used as can the OpenOffice/LibreOffice word processing app. The thing is that you need to specifically save as *.txt. You use the “Edit” “find and replace” function.
The OpenOffice/LibreOffice spreadsheet can also be used with the save as “*.csv”
Kitty, your instructions said:
“A suggested usage is to download your FIA file from DNAgedcom;… ”
Searching for the DNAgedcom web site got me the “DNA Tools” website. Is that where I’m supposed to be?
I clicked on the Members tab and selected View Files. There was no FIA file listed, just:
23607_ChromosomeBrowser.csv
23607_Family_Finder_Matches.csv
23607 _ICW.csv
Where should I look for an FIA file?
Thank you.
-Bill Martin Denver, CO
When you download your data from 23andme one of the files is called FIA which stands for Family Inheritance Advanced. That file has all you overlapping DNA segemnets with the people you are sharing with
I meant to say the 23andme tab on DNAgedcom which yes is the DNA tools website. The files you listed are from Family Tree DNA and the Chromosome Browser file should work with this tool once you change the headings.
After a few days studying, played around with both your chromosome and segment tools to get the picture I wanted this afternoon. The segment tool actually gave me the pic I wanted, the combined chromosome info to a match shared by myself, my mom, and my mom’s 2 maternal aunts. TY
But now, I guess I’m bordering elderly at 56, can’t get the web page to print, can’t copy to print, maybe just too tired after 8 hours of work and need the instructions simpler so I can save the info somewhere for later use.
TY though, for this tool
Linda
Congratulations on getting the picture you wanted Linda! So sorry that it does not print. That surprised me too when I tried it.
You need to do a screen capture and then paste that into a word document or an image file. If the whole picture fits on your screen then try the print screen button. Pressing the alt key and the print screen at the same time saves the current screen image to your clipboard also. If you google “screen capture” you may find something to use with your computer and browser. I use Fireshot with Firefox for this purpose.
If you want to save the image for later review then why not save it to your computer?
You can do that by right clicking on the page and click the save page as web page or the equivalent in your browser. If you have more than 20 relatives shown then you also need the zip file of striped images which is linked to from the bottom of the page – http://kittymunson.com/dna/stripeff99cc.zip
Great tool! I’m having a slight issue with some folks not showing up on the graphic. I have a file with 40 people. Everything is labeled correctly but some of the people listed in the csv file and also listed in the segment “key” don’t show up as bars on the chromosome graphic. Most seem to be left off at random and they are not hidden under any others. Even after re-ordering the file, the most that would show a bar was 32 people.
Send the CSV to kitty at kittycooper.com
Lisa, I think you forgot to check the box that says your data uses decimal numbers. When I checked that your csv worked fine.
Because I mix the two myself, it figures out when it sees a period that that line is decimal. The problem comes when there is no period and it is decimal! So for example your cousin at 140 on chromsome 2 had no decimal so that became too small a segment to show up.
If you forcefully make that column a format with one or two decimal places that would also solve the problem.
Thanks!
Pingback: What Else Can I Do With My DNA Test Results?
Mine does not look like yours at all. Its jumbled and very hard to read. Yours has a nice column down the side, mine has names at the bottom of the screen and a bunch of very small colored/striped bars at the top. Above that is what appears to be two chromosome bars with numbers underneath, the numbers are overwritten by other numbers and are far too hard to read.
Try sorting by segment size so the 40 people with the longest DNA segment matches to you are the ones that get graphed. If that does not work send a screen shot and the CSV to kitty at this website name without the blog. in front
Kitty,
I just noticed a quirk that may affect those who, like me, have Irish families. If you put an apostrophe in your match’s surname, like O’Sullivan, it disables the hover function. If you remove the apostrophe it works just fine.
Thanks for this great tool. Hope it comes to Gedmatch soon.
Jim
Hi Kitty. I came across this tool and it looks interesting. What do you do with the genetic information that is uploaded to your site? As you know, sharing personal genetic information can be quite a dicey subject. Is a stand-alone version of your program available for download?
Thank you!
Trevor
Trevor –
I do not keep these files. They are stored as temp files on my server and get deleted soon after being used to create these images. If you have PHP available on your computer, then you could use the version stand alone. Email me privately kitty at kittycooper dot com if you want that.
– Kitty
Kitty,
Among other things, I use your overlapping segment mapper to track which of my ancestors’ countries of origin are associated with a given segment of my genome. I do this using 23andMe’s Countries of Ancestry Matches (aka Ancestry Finder). I only record a match if the person has indicated that all four of their grandparents were born in the same country. Sounds good in theory, right?
My father’s ancestors are all from Sicily, and my mother’s ancestor’s are from England, Ireland, Germany, Netherlands, and Austria. Below are my matches from 23andMe for a particular segment (approximately starting at base pair 28 million and ending at base pair 37 million) on the fourth pair of chromosomes:
Italy 4 26.4 31.4
United Kingdom 4 28.7 34.4
United Kingdom 4 29.2 34.3
Ireland 4 29.3 36.1
United Kingdom 4 29.3 34.4
Italy 4 29.5 34.8
Germany 4 29.7 34.5
Italy 4 29.7 34.5
Germany 4 29.9 35.9
Germany 4 29.9 35.5
Ireland 4 30 35.3
Netherlands 4 30.9 36.3
Germany 4 31.2 36.6
Ireland 4 31.3 36.6
Again, the country indicated is supposed to be the country of origin of all four grandparents according to the customer. What could account for the variety we see above? Shouldn’t we expect to see only two different countries for a given segment: one being my father’s ancestors’ country of origin (Italy), and the other being one of the countries of origin of my mother’s ancestors? Am I simply unable to rely on 23andMe customers accurately identifying the birth countries of their grandparents? Numerous other customers matching me on this segment list yet other countries from which none of my known ancestors originate. Mobility between European countries can’t account for all the various countries of origin we are seeing, can it?
Jim
Jim –
Nice creative use of my mapper. Yes you can make a chromosome map of anything with these tools.
My latest in progress effort is a specific ancestor chromosome mapper … send in several spreadsheets and it will find a specific name in the MRCA column and show just those segments (and who from) thus mapping the DNA from that ancestor.
Ancestry composition is far from perfected. People in Southern Germany often look like Italians in their DNA. Netherlands, Ireland, Germany, and the UK all have plenty of DNA from the tribes of the Angles and Saxons plus some Viking so can look very alike … plus there was far more within Europe immigration a few hundred years ago than you might realize.
Also see http://www.legalgenealogist.com/blog/2014/05/18/admixture-not-soup-yet/
and try some of the Admix tools at gedmatch
Kitty
Can I use raw data from ancestry.com on your segment mapper?
Sharon –
No you cannot. The mapper is meant to be used when you have a spreadsheet of the overapping segments for your matches. Ancestry data has to be uploaded to family tree DNA or GEDmatch in order for you to see those overlaps.
I discussed this in this post about GEDmatch for my ancestry cousins: http://blog.kittycooper.com/2014/09/using-gedmatch-with-an-ancestry-kit-the-gedmatch-chromosome-browser/
Also here is my post about uploading your ancestry data to family tree DNA where you get a free look but then need to pay to get the real thing http://blog.kittycooper.com/2014/10/free-transfer-of-dna-data-from-ancestry-to-family-tree-dna/
Kitty,
Would it be possible to use this to show how about 40 different people who share descent from one couple match each other? Sometimes there are as many as five or six individuals who share overlapping segments and sometimes only three. (There are many other matches between two individuals, but I would like to show only triangulating matches.) I can’t figure out how to show a picture of this. (Except by using graph paper and colored pencils and then taking a picture.)
The segment mapper can be used to map anything you can make a CSV of.
I used it to map the DNA from an ancestral couple, my gggrandparents here:
http://blog.kittycooper.com/2014/08/mapping-an-ancestral-couple-a-backwards-use-of-my-segment-mapper/
And then did not like the result so improved it here
http://blog.kittycooper.com/2014/09/segment-mapper-tool-improvements-another-wold-dna-map/
Plus on my back burner is making a version of segment mapper that will automate this better, by taking multiple CSVs and map only the entries with the specified MRCA
Kitty, thank you so much for creating this. Would it be possible to make the colors “progress” as they do in a rainbow or the color spectrum, i.e., red, orange, yellow, green, blue, indigo, violet? It would help to “see” the color as a person (red = 1st person, orange = 2nd person) and not have to reference back to the color key.
Bonnie –
You are welcome.
The colors were selected to be visible to the color blind; I had such a person working with me to test this. But you can override the default colors by adding a column called color to your spreadsheet and specifying the color you want by name or hex code. See the documentation above about the color column.
Hi Kitty,
I’ll be buying you a glass of wine for all your help over the past few days!
I’m am now looking into very distant ancestry with someone, to assist me with possible ethnic origins. I match a friend, his parents, and his nephew all on multiple segments, but on chromosome 3, we all pretty much overlap at the same start and stop points, but for a very small length (3cM or less), all well above 500 snps:
Friend 46million – 53million
Mom 46 – 53
Dad. 46 – 53
Neph 46 – 54
Is this situation indicative of a probable common ancestor within the last 200 years or so?
That is too small, it is a likely population specific segment (e.g. Italian or whatever the ancestry is)
I was able to make this work just fine two weeks ago. I’ve been able to identify two new relatives that I was hoping to add to the mix and now it’s stopped working for me. I do have a mac, but have been able to have the csv work. If anything, it will only make one line one a single chromosome nothing else.
Any recent changes or thoughts?
No recent changes or thoughts. Double check the common problems section. Likely because you are on a Mac.
If you cannot resolve this, send me the file to kitty at this website url without the “blog.” part
Hi Kitty,
Along with all the others, I thank you for creating this tool. I am still fairly new to all of this, and it is clear for me to understand.
Today I tried to do a 40 person match–it looked great! I was so excited, then I saved it as a complete web page, I downloaded the strip zip file, extracted it, and Nothing! Maybe I went too far-I put the zip folder in the web page complete folder, extracted it to same, removed the zip file leaving the extracted .gif files. Should I have made another folder within the first and renamed it to ? I do not know anything about programming, so I am probably misunderstanding your directions. Can you help?
Thank you,
Kimmer
Apparently the way it saves as a whole webpage has changed. It might be easiest to just take a screenshot but that does lose the mous overs.
Try to save it from a different browser. If that does not work then you have to edit the saved html file in a plain text editor and replace every " with ” – that is every ampersand followed by the word quot followed by a semi-colon.
Or do a view source of the original page and then copy that source to a file and name it with a .html extension
Hi Kitty – glad I found this! I got a 23andme DNA analysis and a relatives spreadsheet showing shared chromosomes, etc. I am certain about my mother’s father’s line because many of those relatives are also on the MyHeritage family tree I created with the known facts of his lineage. However, my mother was adopted by him and his wife and my (half-) sister Aimee and I have no info about her mother’s mother. So, I’m trying to do some amateur sleuthing by exchanging trees with other relatives, hoping that places and dates will offer hints. My question is: when other relatives and I share the X chromosome, what does that mean? Does that help me determine which side we’re related on? And, btw, I am ordering a few things on Amazon Prime and will use your link as I do appreciate your time and expertise. Thanks so much! Sally
Sally, thanks for using my Amazon link! I really appreciate it.
I recommend you join DNA detectives at Facebook and read the methodology at DNAadoption.com – there is also a yahoo mailing list for the DNA adoption folk.
As to the X, since you are a woman, you get an X from each parent so the X is less helpful. Also a female X match below 10cM is false more often than on other chromosomes.
Can you test your father so you can eliminate his matches? And his X matches?