How do you use autosomal DNA testing to enhance your genealogical research without having to take a PhD level course? This is a question several of my cousins have asked me, so here is my attempt to answer.
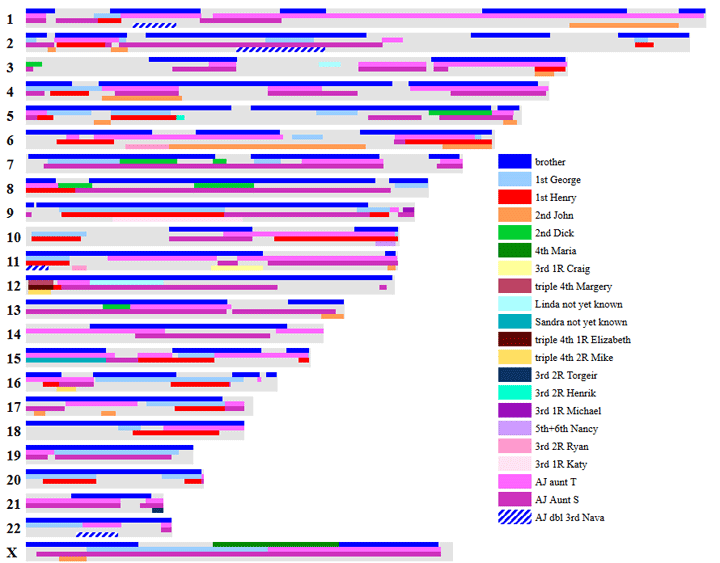
- First of all, get as many relatives to test as you can. The more data you have, the easier it is to make useful comparisons and sort new DNA relatives into their related family lines. The closer family members shown in my chromosome map above are all cousins I convinced to test.
- Secondly, make sure you know how to use a spreadsheet: sorting, deleting rows, inserting columns; you only need to know the easy stuff. [UPDATE: click here for my post with a youtube video teaching that easy spreadsheet stuff]
- Third, check your understanding of how DNA works. Perhaps read my basics page – http://blog.kittycooper.com/2013/04/the-basics-at-23andme/ – and follow up with whichever lessons, books or videos appeal to you among those that I suggest or that you find listed in the ISOGG wiki – http://www.isogg.org/wiki/Autosomal_DNA
Also lots of beginner questions are answered in the FAQ I keep for the DNA-NEWBIES mailing list on yahoo; a copy is on this blog – http://blog.kittycooper.com/dna-testing/newbie-faq/ – so check there when something is confusing. - Fourth, bookmark a page with an explanation of all the acronyms – this is a good one: http://www.isogg.org/wiki/Abbreviations – a key term is cM. You do not need to understand the definition of a centimorgan (cM), hardly anyone does; just accept that it is the best measure of the importance of a DNA match, the larger the better.
Now to the practical application of all this, using shared DNA segment data to find relatives, preferably those 3rd and 4th cousins that your family no longer knows of. Click on success stories on this blog to read about some of the cousins I have found with DNA.
When people are shown as matches to you or your relatives, the testing company will make a guess as to how closely related they are. Beyond 3rd cousins, it is not possible for them to make an accurate estimate because of the random nature of DNA inheritance.
If the expected relationship ends in distant cousin, the common ancestor is likely too far back to find, unless both of you have deep trees (on GEDmatch this would apply to anyone greater than 5 or even 4.5 generations distant). So I recommend only contacting the more distant folk when you see a common surname or common locality. The reason is that once you get past 3rd cousins, the amount of DNA you share with relatives can be fairly random or even none due to the vagaries of DNA inheritance.
See the chart and tables at the ISOGG wiki for the expected amount of DNA shared with relatives
http://www.isogg.org/wiki/Autosomal_DNA_statistics#Simple_mathematical_average_of_sharing
To find possible relatives you want to look at how much DNA, how many segments, and how many big segments you share with your matches. The most promising matches are those with more than one segment and preferably at least one of those segments larger than 10cM. Third cousins will usually match you with one or more large segments but 10% of the time they will not match you at all and about half your fourth cousins will share no DNA with you unless you come from an endogamous group. If your ancestors were endogamous, raise the guidelines above and read my post on Ashkenazi DNA.
Another problem is that sometimes when you share two good sized segments (or one very large one which could be two next to each other), they come from two different ancestors, so the close cousin prediction is wrong.
So how do you find the DNA segments you share with a relative? Well you cannot find them if you tested at Ancestry.com. Ancestry
testers must upload to GEDmatch to see this information.
- At 23andme you go to Tools, DNA relatives, DNA. See http://blog.kittycooper.com/2015/11/initial-report-on-the-new-23andme-for-the-more-advanced-users/ (but if you still have the old version you go to ancestry tools, family inheritance advanced: see http://blog.kittycooper.com/2013/04/the-basics-at-23andme/ )
- At Family Tree DNA. you use the chromosome browser, see
http://blog.kittycooper.com/dna-testing/family-tree-dna-basics/ - At GEDmatch you use the one-to-one see
http://blog.kittycooper.com/2014/10/using-gedmatch-for-my-ancestry-com-cousins
One proviso, if person A matches you at the same place as person B, they do not necessarily match each each other. That is because every chromosome is a pair and the tests only know the spot where you match not which of the two chromosomes it is on: the one from your mom or the one from your dad. Even worse sometimes a person listed as a match is not really matching you at all, but rather matching bits from each parent which looks like a match to the computers. See this post for more on false matches: http://blog.kittycooper.com/2014/10/when-is-a-dna-segment-match-a-real-match-ibd-or-ibs-or-ibc/
Triangulation, when person matches you and another at the same location, is how you confirm a real match. Having a parent and child tested can really help with this. For further reading on triangulation try this blog post: http://blog.kittycooper.com/2015/02/triangulation-proving-a-common-ancestor/
Here is my suggested further reading on finding relatives with DNA: http://blog.kittycooper.com/2013/01/finding-distant-relatives-with-autosomal-dna-testing/
Also you can click the tag DNA spreadsheets to go even deeper into that topic. Plus I have many tools including my own listed on my tools page. The picture above is from my segment mapper tool; I used the track column to move the matches around so it looked better.
If you are an adoptee please go to DNAadoption.com and check out their methodology.
Here are some statistics on my own matches with the X removed (since it is so different between the sexes) . These are some of the matches shown in the chromosome picture above. I do have a 4th cousin once removed (maria) who I share 33.6 cM on the X with but no autosomal DNA. I know it is a good match not just from the size but because my Dad has the same match.
| Known Relationship | total cMs > 7 | Longest Block | # of segs > 7 |
| brother | 2809.80 | 175.00 | 52 |
| NORWEGIANS | |||
| first cousin | 934.90 | 71.20 | 35 |
| first cousin | 758.10 | 49.90 | 31 |
| 2nd cousin | 256.90 | 56.90 | 12 |
| 2nd cousin | 169.90 | 32.70 | 10 |
| 3rd | 69.10 | 22.60 | 5 |
| 3rd | 39.24 | 20.10 | 2 |
| 3rd 1R | 18.90 | 18.90 | 1 |
| 3rd 1R | 13.90 | 13.90 | 1 |
| 3rd 2R | 28.58 | 18.90 | 2 |
| 3rd 2R | 19.80 | 12.00 | 2 |
| triple 4th | 28.70 | 19.30 | 2 |
| Triple 4th 1R | 27.90 | 19.10 | 2 |
| 4th | 0.00 | 5.80 | 1 |
| 5th + half 6th | 17.57 | 17.57 | 1 |
| ASHKENAZI | |||
| aunt | 1646.80 | 172.00 | 40 |
| aunt | 1468.10 | 187.00 | 41 |
| 1st cousin | 821.00 | 142.20 | 26 |
| Half dbl 3rd cousin | 93.60 | 30.70 | 5 |
Those of you curious about what I mean by a half double 3rd cousin, we share 3 gg-grandparents; my g-grandmother fixed up her sister with her husband’s half brother.
AWESOME!!!! I’m sharing!!
Thanks for this informative post. Now if I could just get some of those matches to respond! One thing I started doing on 23andme is putting a person who is sharing genomes with me as one of my parents on my family tree. Then, even if you’re not sharing genomes, 23andme will list your DNA relatives as a “P” (paternal) or “M” (maternal) next to everyone’s name who matches the person you put as your parent.
Nice post, and very informative. I’m bookmarking it for reference. I have a gg-grandfather who may have been adopted (or taken in). I’m getting DNA samples from a direct male descendant of his (may be) adopted father, and from documented descendants of his (possibly) biological father. Beyond that I have DNA results from myself, my father, my brother, and two cousins. The problem is that the descendants, both adopted and biological, are from different mothers than my gg-grandfather, making the relationships half. Right? Do you think I have enough coverage to make a match one way or the other?
Sorry…it’s my great-grandfather that may have been adopted, not my gg-grandfather. Makes everything one step closer.
Ah, 2nd cousins, that will work very well then
Sounds like it might work out, still half 3rd or 4th cousins do not always share DNA so more people would be better.
But if you have direct male descendants then Y DNA testing is an easier way to prove or disprove the possible relationship, although of course not as fun or as informative for the test taker. Howevr if you autosomal test with 23andme you will get the male haplogroups which may or may not answer some questions (different haplogourps would disprove but the same haplogroup is not enough information to prove)
I am not sure I would say that cM is a measure of significance of a match. The word significance is an official statistical term, that implies that some statistical test has been applied, and a number is given to how likely two measurements are similar for example. As a geneticist, centimorgans has been used as a measure of distance between two genes, determined by frequency of crossing over. I am not sure about this but I would guess that cM applied here refers to the size of the DNA match. That is, lets say, my DNA matches 20 cM of yours. But that does not imply significance to the match in statistical terems.
Paul I am not using the term significant in a statistical sense. My mission is to present difficult concepts in plain English using as little of the jargon as possible. The online dictionary defines significant as “sufficiently great or important to be worthy of attention; noteworthy” and that is the sense I am using it in.
The problem is that newcomers to DNA testing get hung up on the definition of cM as a probability and are confused by its use as a size so I am suggesting they ignore all that and get that it is the number to look at to judge the importance of a match.
OK I decided to change “significance” to “importance” so as not to confuse any statisticians in the audience 🙂 Thanks for pointing this out.
I was just wondering how big a factor the fact that my paternal grandmother and her sister married brothers would be.
Olanda, since your grandmother and her sister married brothers, you will have roughly double the expected DNA with any cousins descended from your grandmother’s sister. So double the numbers on the ISOGG chart (link is in the article) when looking at their DNA results compared to yours.
Pingback: Noli Irritare Leones » The Dress, and other blogging and articles about genetics (mostly)
Is there a program, which builds a putative pedigree based on the shared fragments from my DNA matches? Matrix function is good, but building possible pedigree trees from the cvs files will be better. Thanks
Konstantin –
Autosomal DNA is just not accurate enough for such a program to work without much manual intervention. For example, a grandparent and an aunt/uncle look the same in DNA as compared to you, see http://www.isogg.org/wiki/Autosomal_DNA_statistics#Simple_mathematical_average_of_sharing
I occasionally look at your blog, but most of the time it is just too confusing. That said, I’m biting the bullet here in trying to force an understanding. For starters, my focus is on identifying ancestors of an adoptee and trying to use the DNAadoption methodology. I’ve found a few very distant matches through tirangulation. So, following your above points, the only family relatives tested are myself, at 23andme and FTDNA, and my brother with AncestryDNA, all results sent to GEDmatch. We are 69 and 70, there’s no one older than us alive to test. Second, the dreaded spread sheet. With thousand of possible matches and thousands of persons in family trees, I can easily appreciate the need for an organized approach. I have looked at a few tools offered by AncestryDNA, and some of the works ones, but can’t figure them out. Someone helped me make a spreadsheet of my FTDNA matches, but haven’t found that useful. Isn’t there a really simple spreadsheet that assumes the user does not know what they are doing?
Just sign me “Stuck on Stupid”
Christine
Have you taken the class at DNAadoption? that might help you alot. This post of mine might be useful as to why make a spreadsheet
http://blog.kittycooper.com/2012/11/making-a-spreadsheet-of-your-dna-matches/
For help on spreadsheets …
Lynda.com has lots of helpful tech courses, here are the ones in spreadsheets:
http://www.lynda.com/Spreadsheets-training-tutorials/1367-0.html
Also try googling spreadsheets for dummies and see what you find
Ok now that I tested at ancestry.com. lol what is my next step. Should I treat 23 ans me also?
Thanks Ann
Ann –
Sorry for the delay, I have been on vacation.
Your next step would be to load your raw data over to family tree DNA which is $39 (see http://blog.kittycooper.com/2014/10/free-transfer-of-dna-data-from-ancestry-to-family-tree-dna/ ) and GEDmatch which is free. But of course this depends on your objective.
Hello I am new to the whole idea of DNA testing I got my results yesterday from My heritage.com and they say that I am 98.4Ashkenazi Jewish and 1.6Mizrahi Jewish – Iranian/Iraqi.
And now this is where I am confused as I had kind of assumed that it would be more of a 50/50 split with mother and father etc?
But the way I read my results it seems that only shows I side?
Paul –
You get 50% from each parent and it is being reported that they are both fully jewish. Ahskenazi can also be sephardic of Mizrahi, they all look too close in the DNA to really tell them apart. Ancestry composition is still an emerging science and far from completely accurate.
You might want to try the admix calculators at GEDmatch.com where you can upload your MyHeritage results for free. You can also upload them to Family Tree DNA and for $10 in the current sale see their analysis. Both those sites will give you more cousin matches. Neither is likely to do much better with the ancestry composition at this point in time.
Hi, I took the autosomal markers panel 3 test, D9S917 after reading it was the test to take to varify native american lineage. my results were: value 15-17; I took the tat at family tree dna, but they won’t give me an explanation, saying I should ask the person who recomended it. I read this in an article online, which unfortunately I did not save. Can you please tell me what the results mean? thanks.
Robert –
I am sorry, I have never heard of such a test and googling did not offer any answers. If you find the article, by all means post the URL
You might look at this for determining native american ancestry:
https://isogg.org/wiki/Native_American_Ancestry_Finder