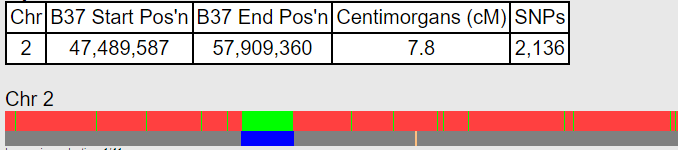
Segment sizes matter for predicting the closeness of a relationship. That is because inherited DNA chunks get divided up with each generation’s recombination, becoming smaller over time. It has often puzzled me that none of the DNA relationship calculators take that into account. The total centimorgans (cM) can be similar for lots of different relationships. Click here for my post on telling half siblings apart from aunt/uncle/niece/nephew relationships by using segment sizes.
Click here for Ancestry’s explanation of how they predict relationships and here for the article at 23andme. Neither mentions segment sizes.
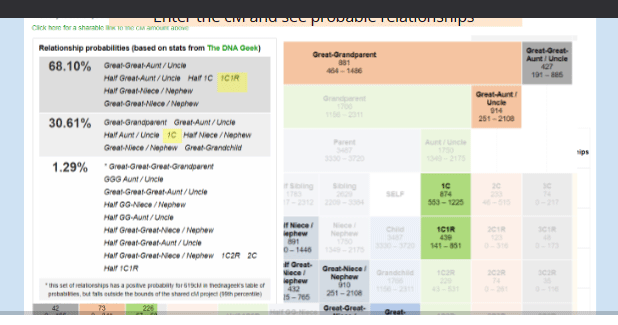
My favorite calculator has long been the one at DNA Painter where the table is based on Blaine Bettinger’s collected data and the probabilities are by Leah Larkin based on the Ancestry white paper. It can work from a percentage or a cM value and lights up just the possible relationships in its table. Clicking on any table entry gets a histogram of frequencies. I have slides showing how nicely it works, done for a recent talk about 3rd party DNA tools for the SCGS jamboree. Click the image above to see those. That video should be available to attendees from the web site genealogyjamboree.com
Recently a calculator was published at DNA-SCI which includes segment sizes by asking for the number of segments. I have used it for a number of closer relationships and have been very impressed with its results.
Continue reading