So what is the impact of MyHeritage doing the family trees for 23andme? We can all agree that the previous family trees at 23andme were difficult to use. So this has to be an improvement, right? Maybe not.
To try it, I connected my own 23andme account to my existing MyHeritage tree and then looked at it from my aunt’s account, after logging out of MyHeritage. It was hard to find it on my profile. All the way at the very bottom were the words Family Tree next to which were the linked words View Kitty Cooper’s Family Tree. When I clicked on it I saw this:
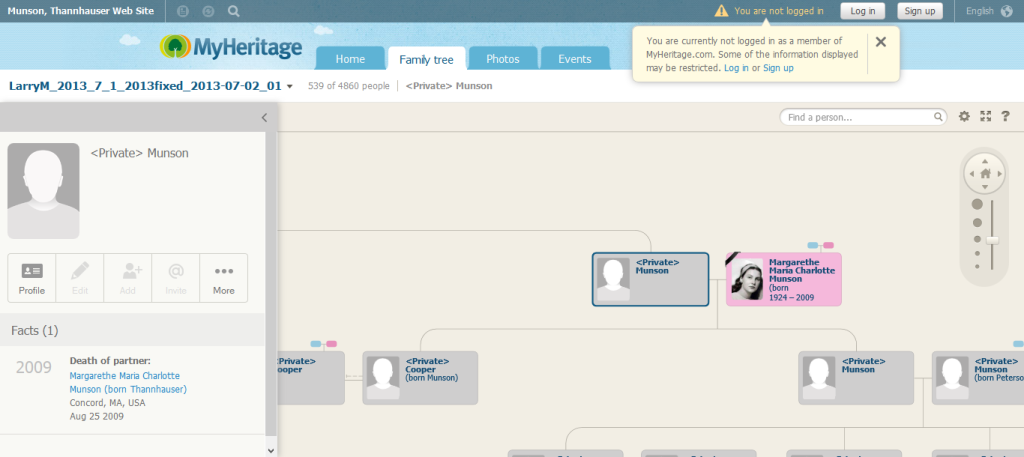
Oops. My Dad is set as the home person. Maybe Google can help me figure out how to change that. Clicking on the image above will take you to my tree so you can judge for yourself how well you like it.
MyHeritage is an excellent site in many ways, but to take advantage of the record matching you have to buy a premium membership. Also if you want more than 250 people in your tree, payment is required after those first free 6 months for 23andme folk. The premium or data membership gives access to many European records not on ancestry and also some newspaper articles I have not seen elsewhere.
For me, the biggest issue with MyHeritage is having no pedigree view on the family tree. When generation after generation had eight or more children, their tree view is very unwieldy.
In the rest of this post I will walk you step by step through setting up your 23andme tree at MyHeritage which you must do by May 1, 2015 to take advantage of the offer.
Continue reading