A team of Penn State researchers has made a map of human chromosomes that shows the areas where mutations are more and less frequent; in their words “mutationally hot and cold regions.” However I found their diagram extremely difficult to understand. It took me quite a while to figure out the areas that are hot and cold for the SNPs that genetic genealogists are interested in. So I redid their image, removing the color for microsatellite repeat alterations, and changed the colors a little to be more in tune with hot and cold for me.
Here is my version:
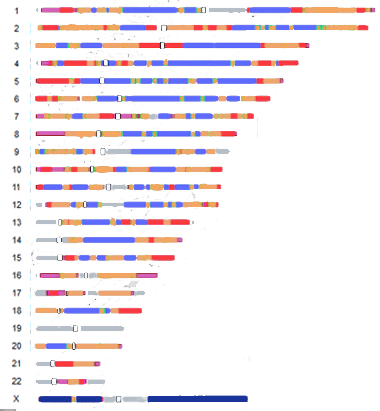 |
Gray presumably are the areas not done and white outlined with black shows the centromeres. The one place with the least mutation is the X chromosome. |
No guarantees that my reinterpretation of the graph by Kateryna Makova and Francesca Chiaromonte is correct!
So what does this mean for genetic genealogists? Well I would think that matches in the pink or red areas (hottest and hot) are going to be more recent and perhaps findable. Personally that blue area of chromosome 9 is where my Dad and I match many many people with no possible ancestral connection to us in the last 500 or so years! None are from his ancestral home area (Southen Norway) and most of his lines are documented pretty far back.
Here is the full article:
http://medicalxpress.com/news/2013-08-characterization-human-genome-mutability-catalyzes.html#nRlv
Warning it uses highly technical terminology
Kitty–this is very helpful. Could you provide the numerical data underlying the map? I’d like to copy it into my own chromosome maps for comparison.
Thanks,
Jim
Jim you would need to look at the original article for that data.
Fascinating! Compared with some of my more remote MRCAs and they fall in line with your chart. It gives me a little more confidence that we might be on the right track.
I tested at 23andMe earlier this year and have been comparing genomes for a while. I have over 300 people that I share/compare genomes with. And it always seemed to me that some areas had more matches than others. This article confirms my suspicions. It seemed like rather large segments seem to pass down through the generations and this makes matches more noticeable. One group of people in particular are a good example of this. I match at least 4 people in the blue area of chromosome 18. The match is between 15 and cM in size between all of us. And it is the only matching segment that we have. I was able to find a common relative with one of them that dated back to 300 years ago. Does that mean that we all share this common ancestor or are you saying that the connection could even be further back?
Most of us have at least one spot in a blue area with many matches that must be far back in time.
The connection could easily be the ancestor you found or further back for the others. This article figures out mathematically that 10 generations is about the limit for detectable DNA inheritance:
http://www.genetic-inference.co.uk/blog/2009/11/how-many-ancestors-share-our-dna/
I am the one who posted the question about the multiple Chr 18 matches.
Would you be so kind as to send me as much as you can of the details of your matches on Chr 18, so that I can see whether these indeed match with the half dozen that I have? Does 23andme give you the positions along the chromosome in mB? I have no experience with it, being an FTDNA person.
and I am intrigued by the data showing a family connection going back 300 years or ten generations. This would be quite mysterious and I could take the matter up with some professional geneticists who work on recombination.
I have checked with an author of the hot spots mutation paper cited earlier in this string. She agrees that this work need have no connection with recombination jungles (as seems to be the preferred term in the case of recombinations)
w
See this article http://blog.kittycooper.com/2013/02/largeibdsegmen/
which does the math on single segment matches showing that they can be far back. Certainly in my Norwegian families where 9 children were normal the odds increase.
Also 23andme rounds off the positions which makes sense as the borders are fuzzy anyway. Sending you chromosome 18 data via email
My ears perked up when I saw this post mentioned on an Ancestry board. (I’m waiting on my Ancestry results)
I have 12 matches over the centromere on Ch 19, which isn’t colored at all~ why is that? And a lot of my matches seem to be from Denmark. (I’m adopted).
Also, do you have a post about the significance of matching across the centromere? Thanks 🙂
Hi Callie –
Well it was not my experiment or data, I just reinterpreted the diagram for us genetic genealogists. I assume the gray areas are neither slow nor fast, but it could just be that they did not have enough data. I reread the article and still cannot tell.
The centromere is one of the hottest recombination points there is. Therefore I would consider a match across it to be two separate segments. The FAQ at familytreeDNA has the location of the centromeres
http://www.familytreedna.com/faq/answers.aspx?id=17#610
So what I do is subtract that out of the middle and list the overlapping segment as the two resulting ones in my master spreadsheet.
Last but not least since you are adopted have you read this in my newbie FAq? http://blog.kittycooper.com/dna-testing/newbie-faq/#__RefHeading__792_704084449
I am in those groups 🙂 I feel like I’m swimming in so much information. Hard to sort it out. It’s like the answers are there but just beyond my reach.
It really takes time to absorb it all. Keep reading and listening and be patient with yourself.
The adoption list regularly gives a class in their methodology and also I recommend Kelly’s Basic Genetic Genealogy lessons and my FAQ. I also took a few genetics classes at coursera but those get pretty deep!
Part of the problem is the amorphous nature of autosomal DNA. It gets more and more random the further away you get from each ancestor as to how much DNA you have from them. For example I have one jewish grandparent but I got 27% ashkenazi from him, not the expected 25%, while my brother only got 20% …
Kitty, these are regions of high or low mutability.
I think most of us having the problem of many matches with putatively unrelated people are concerned rather about high or low RECOMBINATION.
This is quite a different issue.
Do you agree?
wilfo
Yes those are different issues but mutability is the more important for matching relatives. What makes us different from each other is the combined mutations we inherit from our ancestors. The autosomal DNA tests we are all using are looking at SNPs prone to mutation NOT our whole genome. Mutation usually means that an A becomes a G not a Z, in other words the 4 building blocks (AG,CT) are the same but the ordering might change. So I am interested in spots where change is more and less frequent. My thinking is that the cold areas on the graph are places where I am more likely to have a match too far back in time to find the relationship.
Here is a good article about mutations: http://evolution.berkeley.edu/evolibrary/article/0_0_0/mutations_03
As to recombination, there are “hot spots” where it happens more often. DNA seems to recombine in large chunks and the boundaries are often those hot spots. I think that is why the start and stop points for segment matches are often the same, or close, even when they are from different sides. Article on hot spots – http://en.wikipedia.org/wiki/Recombination_hotspot and a much more technical one: http://www.plosbiology.org/article/info:doi/10.1371/journal.pbio.0020190
I have a large grouping of matches on number 12 with Italian ancestry, and rate 29% Italian ancestry with no known or confirmed recent Italian relatives. Is that possible?
Thank you for any light you can shine on this for me!
Kathy
Ancestry composition is far from an exact science. Have you looked at the calculators at GEDmatch?
The Romans spread all over the Mediterranean and through France and Spain, leaving ‘Italian’ genes everywhere ….
How large is this match area? It could be a very old piece of DNA.
23andme gives me 29%. Ancestry groups the result Italy/Greece but say may include surroundinf countries. That average is 36%. Is GEDmatch a specific website? According to 23andMe, the match area is 22.5cm. And a different segment below 13.9cm. Those two seem to be where my high % is originating.
Thanks so much!
Kathy
Kitty,
So I uploaded my DNA to GED Match but I am feeling very under qualified to interpret the results. I think if I understood how (the basis) the results are grouped and their meaning, I would be able to make some sense of the data. Is the data results based on how my DNA matches or compares to other individuals from various regions, and it varies based on the number of participants? Or is the data stagnant and does not change regardless of the number of particpants and where they state their relatives are from?
Thank you for your help with this!
Kathy
See if my slide show helps,
http://slides.com/kittycooper/gedmatch#/5
ancestry composition is far from an exact science
each calculator at GEDmatch links to the citizen scientist page which explains how the results are calculated
Kitty,
Thank you – yes I used your tutorial when I first uploaded and generated results. I will have to do some heavy reading and see if I can figure it all out.
I guess my original question is still now answered – and perhaps will never be. Is it possible to have a large % calculation of Italian ancestry (29%) from a great-grandparent? I am trying to solve this nagging mystery. Thanks so much for your patience with this non-scientist!!
Best to you,
Kathy
Meant to phrase it – my original question is still unanswered..
Kathy
kathy,
What is your expected ancestry? There could be many explanations. A grandparent that is not the one you expect … 2 Italian g-grandparents … But again, you cannot take ancestry composition all that seriously. It is far from a proven science. Italian could also be jewish. It is in my family but we are talking 5 or 6 % in our case
Kitty,
I have no known recent Italian relatives. I have a great-grandmother who was presumed to be half Cherokee. She was born in 1882 and found orphaned on the Chisholm Trail – presumably during the Trail of Tears. I have no American Indian DNA, according to my test results, however, I have 29% showing as Italian on 23andMe, and 36% showing as Italian/Greek through Ancestry.com DNA. Those are pretty high percentages. Could it be possible, that, combined with a small amount of old DNA and perhaps my great-grandmother, that my percentages could be so high? The only other explanation is either I’m not my parents’ child, or one of them is not their parents’ child. From what ancestry I have researched online, my father’s father’s line can be traced to Germany, his mother’s father’s line can be traced to Germany, his mother’s mother’s line can be traced to Ireland/Scotland. My mother’s line I am having difficulty with, but the most recent surnames (and physical appearances) do not point to Italian heritage, although there may be some speckled in there here and there I suppose. Mainly Irish, English, German.
Trying to solve a little mystery.
Thanks so much!
Kathy
I uploaded my data to GED Match and the Eurogenes K15 produced the following results:
Admix Results (sorted):
# Population Percent
1 North_Sea 25.30
2 Atlantic 17.17
3 West_Asian 13.69
4 West_Med 13.31
5 East_Med 11.50
6 Baltic 9.20
7 Eastern_Euro 5.12
8 Red_Sea 1.86
9 South_Asian 1.65
The MDLP World-22 Oracle results are:
Admix Results (sorted):
# Population Percent
1 Atlantic_Mediterranean_Neolithic 34.98
2 North-East-European 34.05
3 West-Asian 15.51
4 Near_East 8.23
5 North-European-Mesolithic 3.71
6 Indo-Iranian 1.46
7 Melanesian 0.87
8 Indian 0.45
9 South-African 0.42
10 Sub-Saharian 0.3
Both 4 populations produced a number of German, Greek, Armenian, Swedish, Norwegian, and Georgian. I can post that, too, if you would like to see. I don’t know what it all adds up to – other than Heinz-57.
It sounds like that orphaned g-grandmother was Italian or from somewhere mediteranean and just looked Indian. South Germans often have lots of Italian (or general southern european) because they moved north into vacant lands after the plague, among other reasons, so probably your German ancestry is part of that “Italian” designation.
What does K12 show? and K13?
This slide shows the admix for a fully Italian person
http://slides.com/kittycooper/gedmatch#/14
Yes most Americans are very admixed from the various European populations so welcome to that club!
If you are really worried about this, get more family members to test, particularly cousins. Can you test your parents?
Kitty,
That is what I am thinking – that my great-grandmother was actually full-blooded Italian and not Indian. She is on my father’s side – his grandmother. My grandfather was very dark-haired and dark-eyed – darker skin-toned (from what I can tell from black and white photos). He actually looks like he could be Italian. Both of my parents have passed away, however, I do have female cousins from my mother’s sisters. So I am assuming they should be from the same haplogroup as I am – I2 (since our mother’s share the same mother). I will show you my results from K12 and K13.
Thank you!
Kathy
Kitty – here are the K12 and K13 admixture results. Couldn’t get the pie chart to post correctly.
Eurogenes K12 Admixture Proportions
Population
South Asian 0.57%
Caucasus 19.77%
Southwest Asian 4.77%
North Amerindian + Arctic –
Siberian –
Mediterranean 18.62%
East Asian –
West African 0.41%
Volga-Ural 4.04%
South Baltic 9.29%
Western European 21.99%
North Sea 20.54%
Eurogenes K13 Admixture Proportions
Population
North_Atlantic 32.41%
Baltic 16.56%
West_Med 17.48%
West_Asian 14.48%
East_Med 13.62%
Red_Sea 1.74%
South_Asian 1.73%
East_Asian 0.63%
Siberian –
Amerindian –
Oceanian 0.34%
Northeast_African –
Sub-Saharan 0.99%
Best,
Kathy
Get your female cousins on your mom’s side to test so that you can confirm that this mediterranean 29% is coming from your father’s side and mainly from that g-grandmother.
Again, ancestry compostion is far from an exact science.
If you donate $10 to gedmatch you can access the tier 1 utilities and can run the “who matches me on which segment” utility so you can see if you have a big stack up on those 2 “Italian” segments see http://blog.kittycooper.com/2014/10/new-utilities-at-gedmatch-tier-1-for-paid-members/
Thank you Kitty! You have been more than helpful and I truly appreciate it. Best regards,
Kathy
I know lots of people have claim they have Native American ancestry and it is often a myth. I guess my family is no different. I have been told my maternal great grandmother was part or full Native American. Using the Eurogenes K13 Admixture my 13th chromosomes shows a 21.1 for Native American. Is this number of any significance? Does it mean the probability is higher that I have Native American ancestry. All of my admixture pie charts show a 1-2% Native American
Deborah –
You would likely have inherited around 12% from a great-grandmother so if she was fully native american that is what you would see rather than the 1 or 2% – BUT ancestry prediction is not very accurate yet from any company AND DNA inheritance is pretty random. If she was only 1/4 native american that might be what is showing. Do you also see some Asian, that is how native american is sometimes reported.
That chunk on your chromosome 13 is surely from her
Try the MDLP admix tool as well.