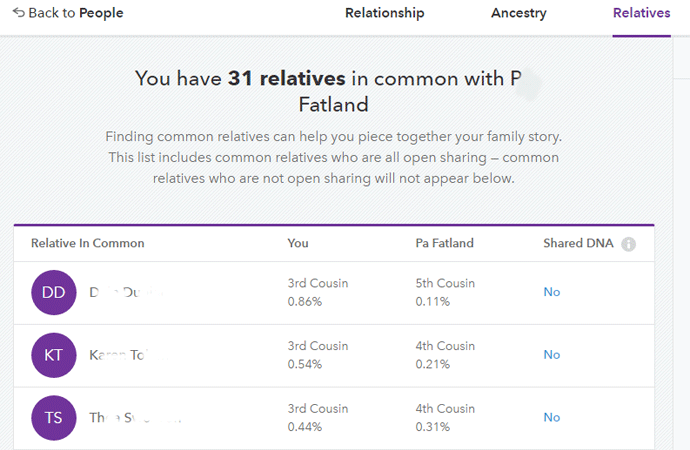
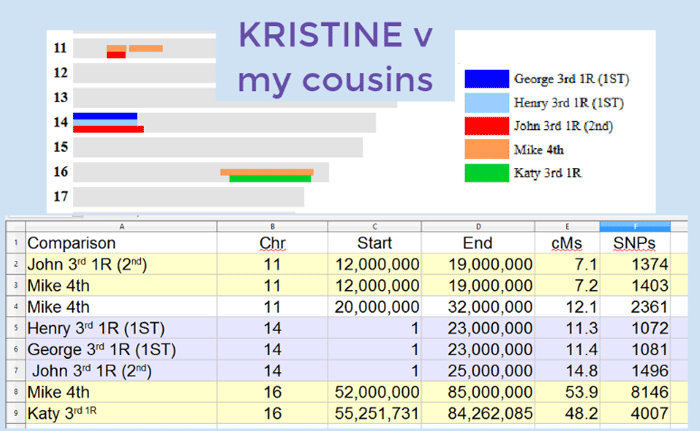
MyHeritage has kept its promises: tree matching, pedigree display, a place for notes, and best of all, a chromosome browser. Plus the cousin matching is finally quite good, at least for your closer cousins, and includes some triangulation.
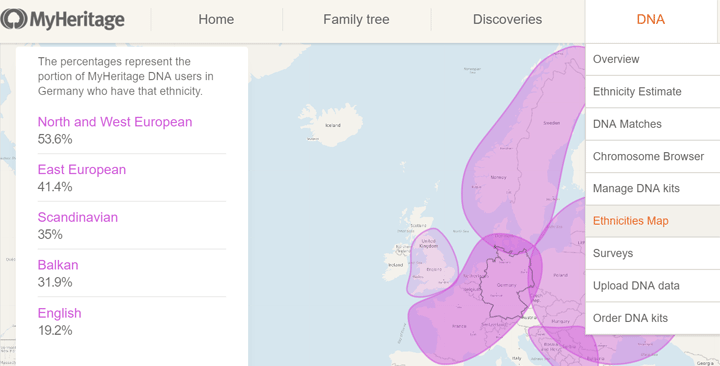
A very nice new feature is the Ethnicities Map, a menu item under DNA, which gives you the common groups for any modern day country you select. Since a question I commonly receive from family members is “Why doesn’t my known German ancestry show up?”, it is great to be able to show them this map:
A picture says it better than telling them that in the DNA, northern Germans look Scandinavian, southern Germans look Italian, eastern Germans look East European, and western Germans look French. My maternal ancestors lived at the crossroads of Europe!
Uploading your results from another DNA testing company is still free at MyHeritage and you get many of the DNA features. Personally I have just a data subscription and a small tree (there is a 250 person limit for unpaid members). In a few weeks I will create an account for a cousin and see if this works as well as it is supposed to for completely free members.
After the recent change, the segment details for my matches to my close family are very similar to what I see on GEDmatch and 23andme, same chromosomes, similar sizes, slightly different boundaries. This is a wonderful improvement!
Since my ancestors are all fairly recent immigrants from Norway and Germany, I was hoping for some international matches when I uploaded my DNA results to MyHeritage last year. In practice, as usual, there were no Germans (testing is not popular there), but plenty of Norwegian cousins that I already knew about, plus a few new distant ones.
However, I did recently get a new close cousin match (1C2R-2C1R), Melissa from New Jersey. I will use her match to investigate the new improved DNA matching.
Continue reading