According to family lore, when their business needed money my great-grandmother Charlotte Langermann Thannhauser, on my German Jewish side, fixed up her sister Lina with her husband’s half brother in order to get Lina’s dowry invested in the family enterprise. Not such an uncommon occurrance. I was able to convince Lina’s great-granddaughter on the straight maternal line to test at 23andme. She is almost doubly my third cousin, sharing DNA from the Thannhauser and Langermann lines but not Engel.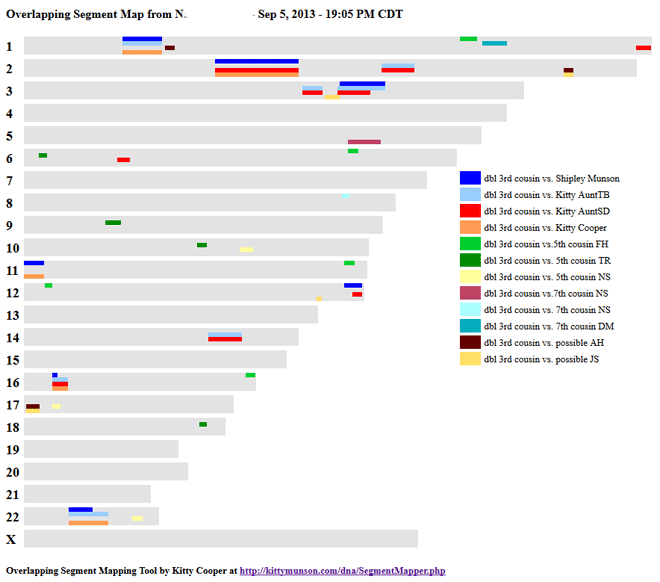
You may ask what this all means. The chart shows places where our chromosomes have a long set of identical SNPs and so that DNA segment is considered a match. A single-nucleotide polymorphism or SNP is a location in your DNA strand which has good variance between people. What an autosomal DNA test does is to sample your DNA by testing SNPs of interest, rather than every base pair. A base pair is the basic unit of DNA; it is made up of two molecules, either an AT or GC – Adenine (A), Cytosine (C), Guanine (G), or Thymine (T). If you were to have your full genome sequenced that would test every base pair.
Because the test cannot determine which of your two chromosomes has which DNA, it looks for long runs of identical SNPs in order to assume we have both inherited the same bit of DNA from a common ancestor. In this case, for me, I know my match with my cousin would be on the set of chromosomes I inherited from my mother’s side.
The chart displays these matches in terms of centMorgans (cMs), which is a measure of the distance based on the frequency of recombination rather than an actual number of SNPs or base pairs. That physical size can vary greatly but tends to be about a million base pairs. Half IBD refers to the fact that the match is on one chromosome only and “Identical By Descent.” Usually only siblings would have any full IBD matches. The page called autosomal images on this blog has a diagram showing my matches with my brother which include some full IBD segments.
By comparing my cousin to my family I can see what DNA we all share in this picture. The numeric details are below.
We now know that my g-grandmother was mtDNA haplogroup K1a1b1a and thus my grandfather’s maternal line back to Regina Guggenheimer was a known ashkenazi haplogroup. I think I was hoping it would be more rare and a help in finding more about her ancestry.
Since I had previously set up a share, I was able to use the ancestry tools > family inheritance advanced to look at the shared DNA with my family which is quite a lot. See the diagram above. It is exciting to me to know which DNA most likely comes from those specific g-grandparents now!
Here are the actual numbers for our overlapping segments. I am orange, my brother is blue and my aunt is yellow in the spreadsheet shown below. The Start and End points are rounded off and are in terms of base pairs (GC/AT) on the chromosome. The genetic distance in cMs is considered the most significant for matching. Typically the larger the cM match, the closer the relationship. Not every base pair is tested, just the ones most likely to be different so that is what is shown in the number of SNPs column.
| Chr | Start point | End point | Genetic distance | # SNPs |
| 1 | 39000000 | 55000000 | 13.6 cM | 2657 |
| 2 | 76000000 | 109000000 | 22.7 cM | 4544 |
| 11 | 1 | 8000000 | 17.2 cM | 2571 |
| 16 | 11000000 | 17000000 | 9.4 cM | 1186 |
| 22 | 17000000 | 32000000 | 30.7 cM | 3416 |
| 1 | 39000000 | 55000000 | 14.2 cM | 2760 |
| 2 | 76000000 | 109000000 | 23.5 cM | 4655 |
| 3 | 125000000 | 143000000 | 17.5 cM | 3294 |
| 11 | 1 | 8000000 | 17.2 cM | 2571 |
| 12 | 126000000 | 133000000 | 21.6 cM | 1987 |
| 16 | 11000000 | 13000000 | 5.0 cM | 712 |
| 22 | 17000000 | 26000000 | 20.4 cM | 1856 |
| 1 | 39000000 | 55000000 | 13.5 cM | 2632 |
| 2 | 142000000 | 155000000 | 10.3 cM | 2176 |
| 3 | 110000000 | 118000000 | 9.8 cM | 1772 |
| 3 | 124000000 | 143000000 | 19.3 cM | 3724 |
| 14 | 72000000 | 85000000 | 14.5 cM | 3148 |
| 16 | 11000000 | 17000000 | 9.1 cM | 1157 |
| 22 | 17000000 | 32000000 | 30.7 cM | 3416 |
I am puzzled by the lack of X chromosome matches though. There is no reason for us not to share X from the Langermann sisters. Perhaps this is just not processed yet. I will try again in a few days and also look more closely when the GEDmatch upload for my 3rd cousin has processed.
Of course since this is Ashkenazi DNA it is possible that some of this comes from elsewhere. My cousin’s father is also Ashkenazi from much further East and AJ DNA is notoriously overlapping with distant connections. So more analysis yet to come. Sadly she has no overlaps with us and my five distant cousins on the Steinhardt/Engelman lines from Floss, although of course she has matching DNA with them. Note that her grandfather, like my grandmother, was non-jewish German but not related that we know of.
All the women in the photo above have mtDNA haplogroup K1a1b1a. Auguste is the mother of Charlotte and Lina, and the young girls are Lina’s daughters.

I was really puzzled by the lack of an X match since our g-grandmother’s were sisters and I have only one male in between. However when I compared the X chromosomes at GEDmatch, I discovered about a 6cM match which corresponds to part of her doubly ashkenazi section on her chromosome level subregional ancestry composition. So the X she got from her mother was fully half from her nonAJ grandad.
I’m having a hard time figuring out the whole cM/SNP relationship for deciding whether or not people are related by IBD or it’s just an IBS match. If you have a family of 8 tested individuals and an “outsider” matches them to one degree or another on the same locations and on multiple chromosomes, do the cMs/snps have to meet a 700cM/700snp rule for a distant cousinship. In my example the “outsider” matches family members in the following way: 5 people in 5.1cM-621snp, matches 2 in 7.8cM-547snp, matches 4 in 4.0cM-999snps, matches 2 in 3.6cM-2141snps, matches 2 in 3cM-571snps, matches 4 in 5.6cM-982snps, matches 3 in 3.3cM-659snps, matches 2 3.6cM-571snps, matches 2 3.0cM-508snps, plus about 15 other low value (3 to 5cM and 500-2000snps) matches that aren’t shared by multiple family members. Are these values all too low to mean anything and just coincidental or could this “outsider” be distantly related to this family?
Linda – If an outsider matches more than one family member for 5+ cM (even less possibly when lots of members match) then it is likely IBD, but it will probably be too far in the past to find unless you have very good paper trails. The cMs are a better indicator then SNPs because a cM is a measure of recombination frequency. See this question in the newbie FAQ for an idea of what is significant: http://blog.kittycooper.com/dna-testing/newbie-faq/#__RefHeading__471_57540906 as well as a link to a good article on the subject
Thank you for your response, and no, I do not have a good paper trail but your answer and the resources available on your blog help to clarify my confusion! I’m really enjoying reading about your “adventures” in genealogy.
linda
wow—I like the graph—I have a few second cousins marrying in my tree, so it looks like I’m more closely related than we actually are.