Many people have the illusion that if their testing company says a person is a 3rd to 5th cousin they really will be. That is not the case.
The testing companies are just making the best guess they can from the data they have. They do not seem to take segment sizes into account, rather they primarily use total shared DNA measured in centimorgans (cMs) for their relatedness estimates, usually the sum of all matching segments of 5 cM or larger. Close relatives will always share larger chunks with each other and so size does matter here.
Recently I have received numerous questions from people trying to figure out if a new match is a half sibling or a niece or a grandchild. These are hard to tell apart without testing more relatives as they all share about 25% of their DNA with each other. So I decided to collect some detailed statistics on those specific relationships with a google form (click here) that includes total segments and segment sizes for a future blog post [UPDATE as of sept 2017: First round results are written up at https://blog.kittycooper.com/2017/09/the-25-relationship-a-first-look-at-the-data/ ]
The companies predict reasonably well for close family but it is just not possible to be accurate beyond that due to the randomness of DNA inheritance.
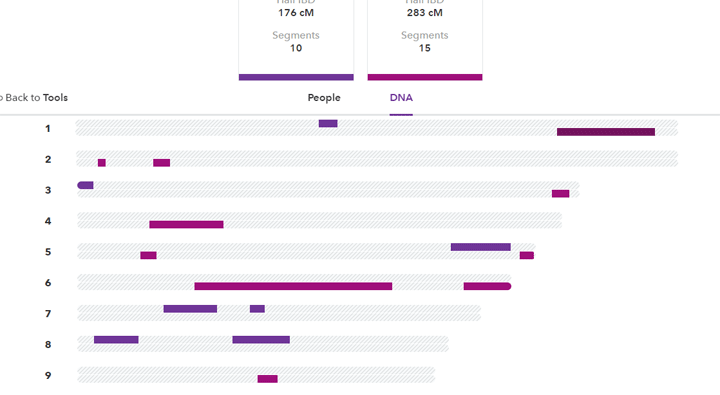
For example, here is a picture from the new 23andme of some of the DNA I share with Dick, a 2nd cousin on Dad’s paternal side so blue, and John, a 2nd cousin on Dad’s maternal side so red.
I share a third again as much DNA with John as I do with Dick, even excluding the 14 cM on the X. The expected amount for a 2nd cousin is 3.125% which is 212.50 cM, right in the middle between these two.
 Checking my brother, I see the same effect – he has 282 cM with John versus 185 with Dick. Not surprisingly, when I look at Dad I find that he shares almost twice as much with John as with Dick. Clearly he just inherited more of the same DNA as John’s mother from their common grandparents. Conversely, he inherited less DNA shared with Dick’s mother from his other grandparents.
Checking my brother, I see the same effect – he has 282 cM with John versus 185 with Dick. Not surprisingly, when I look at Dad I find that he shares almost twice as much with John as with Dick. Clearly he just inherited more of the same DNA as John’s mother from their common grandparents. Conversely, he inherited less DNA shared with Dick’s mother from his other grandparents.
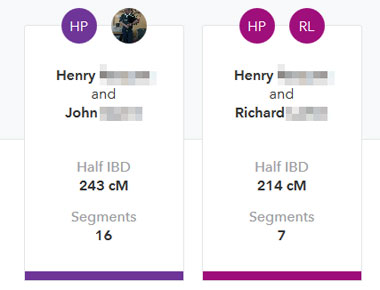
On the left is a comparison of my first cousin Henry with both Dick and John. The amount he shares with each 2nd cousin is practically identical, as long as you subtract the 40 cM that he shares with John on the X from the total shown by 23andme. Amazing how variable DNA inheritance can be among 2nd cousins.
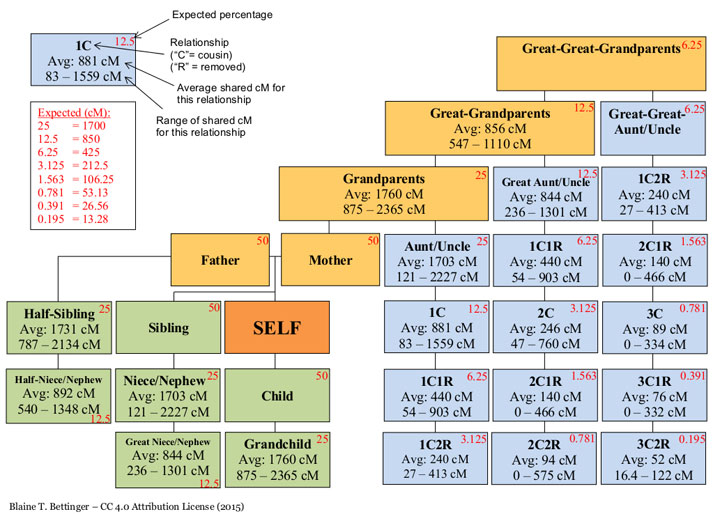
Click here for the ISOGG wiki article on Autosomal DNA statistics which usually includes the current chart from Blaine Bettinger’s shared centimorgan DNA project.