Sometimes I wonder if the interest in genetic genealogy runs in our DNA. I have found many more new cousins with autosomal testing that are descended from my WOLD line than in any other family. Yes they all had lots of children until recently, but so did the Munsons and the Skjolds.
So I decided to make a picture of the HIR (half identical region) DNA segments that I know come from my great-great-grandparents Jørgen and Anna Wold of Drammen, Norway. To do this I made a CSV file with a list of all the segments that are just from those ancestors. I put the first names of the group of matches in the column that would be the MRCA in the usual style segment map. I have to give credit to my distant DNA cousin (on the AJ side) Israel Pickholtz (he blogs too) for this wonderful idea of making a reverse segment map. Below is my picture of Wold DNA created with my DNA segment mapper tool. Click the image to go to the actual html page which will show the centimorgan values and names when you put your mouse on a colored block.
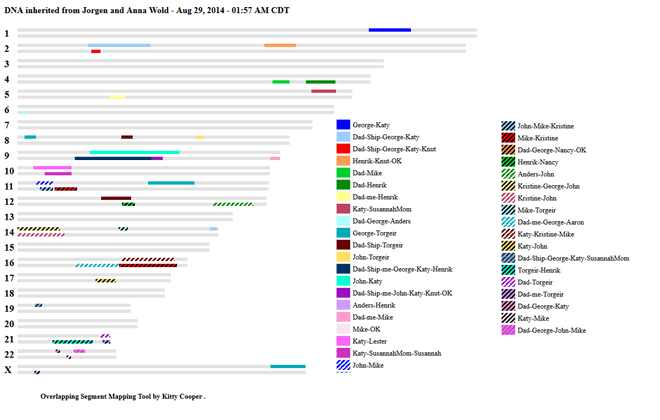 The use of two lines is arbitrary; I could have used three or four. The DNA segments shown are not separated into lines for Anna and those for Jorgen. Where that was possible, I had intended to do it with colors, but did not get to it, next version. Knut, OK, Nancy and Aaron are on Anna’s side while Susannah, her mother, and Lester are on Jorgen’s side only.
The use of two lines is arbitrary; I could have used three or four. The DNA segments shown are not separated into lines for Anna and those for Jorgen. Where that was possible, I had intended to do it with colors, but did not get to it, next version. Knut, OK, Nancy and Aaron are on Anna’s side while Susannah, her mother, and Lester are on Jorgen’s side only.
Segments with only two people are not triangulated and therefore are not proven to be from Anna and Jorgen. I have put them in my chart anyway, because these are such recent ancestors that the DNA is unlikely to be from anyone else. Some are quite large: 25 and 30 cM.
Click here for the definition of triangulation at the ISOGG wiki which states “If you have at least three people with the common ancestor matching on the same segment then you can infer that the segment came from that ancestor.” Because my mother was from a very different population group, when my brother or I have the same match as our Dad, I consider it proven by triangulation. Therefore every segment with three names, even if two are my Dad and me or my brother Ship, is a clear match.
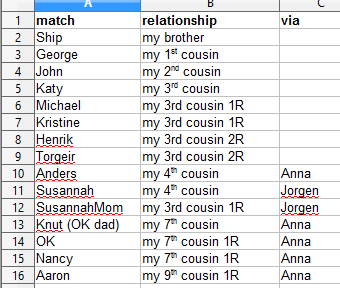 To the left is the list of the people in the chromosome chart, besides Dad and myself, with how they are related.
To the left is the list of the people in the chromosome chart, besides Dad and myself, with how they are related.
Obviously with my close family, I only compare them to 3rd cousins and further out cousins to be sure the DNA is from my WOLD gg-grandparents.
To make the DNA segment picture, I had to be able to compare all our 3rd cousins to each other and to my closer relatives. For the ten of us tested on 23andme, I could just compare five at a time on the FIA page to each cousin. I also used the tool provided at DNAgedcom to download all my 23andme matches in common with my shares for each of my known WOLD line cousins. Plus I used the chromosome browser at GEDmatch for the 3rd cousin who tested only at family tree DNA to see if she matched any of the other cousins. Sue Griffith describes that technique in her latest blog post on triangulating at GEDmatch while that tool is not available. I also used the one-to-one so I could get the number of SNPs. I included a cousin and her mother who are descended from Jørgen’s parents as well as several distant cousins known to be descended from an ancestor of Anna’s. See the my deep triangulation blog post for more about cousins Nancy and Aaron
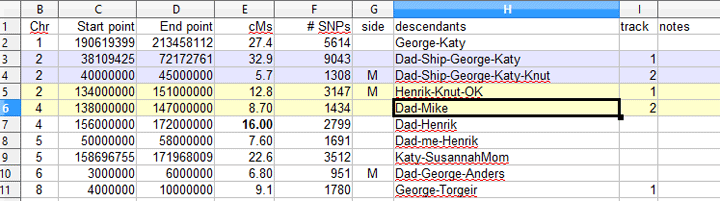
Below is an excerpt from the spreadsheet I used. When there was an overlap but one cousin had a much larger matching segment I put both in the CSV and used a highlight color in the spreadsheet to show that they are the same. Since I used only two tracks (lines) in the chromosome picture, I manually assigned some of the tracks via the “track” column to keep the picture clear. The “side” column is not needed for the segment mapper so that column is ignored. The number of tracks can be set in the form that makes the chart.
Kitty,
I had no idea that when I asked and you responded that it would be so wonderful in so many ways. Thanks again cuzzin for making this awesome tool that so many enjoy!!!!!
Kelly Wheaton
Thanks Kelly, making a tool to create chromosome images from CSV files turns out to be far more flexible than we realized at first.
And of course this is the line you are probably related on as well! Anna had ancestors from Seljord, Telemark and I photographed a bunch of pages from the Bygedebok when last in SLC. So new information to be on my ancestry tree soon! Then I will review our small match again
Anything to get me back into what is important!
Thanks Kitty, this was really interesting to read. Torgeir
Glad you enjoyed it cousin Torgeir. I am not happy with the picture though so I am revamping my segment mapper a bit to improve upon it.
I am amazed that you have such a big piece of your X chromosome (36.5 cM – 2401 base pairs) from Jorgen and Anna which you share with my first cousin George.
You two are both descended from Wold daughters and then their daughters and then their daughters … then comes my cousin and your maternal grandad. A long time for such a large piece to stay together!
I am constructing a reverse chart for a 2x gt grandfather and his two wives. Do I assign the descendants to a maternal/paternal side depending on their direct parent’s sex, which 2x gt grandparent they are descended from, or leave it as unknown?
ZZeh
It is entirely up to you but when I reverse chart an ancestor I use the segment mapper not the ancestor chromosome mapper. The segment mapper has no side parameter. You can specify how many tracks to use, for example just one!