The DNA segment chromosome mapping tool is now released for all to try. This is the current URL:
http://kittymunson.com/dna/ChromosomeMapper.php
and the documentation is here:
http://blog.kittycooper.com/chromosome-mapper/
Thanks to all the testers, I think the bugs have been found and fixed. The next release will have some enhancements so make your suggestions here. So far I expect to add the following features:
- ignore the side column so alternately chose a side and color
- better error reporting and more bullet proof
- add a third option and thus line on the chromosomes for unknown or both
- add a note column that is part of what appears on mouse over with the MRCA
- allow selection of colors
- make a version designed to show overlaps on unknown relatives
This was conceived as a tool to show which ancestor which segment came from by mapping a CSV file listing ancestral DNA segments. Like the pictures Cece Moore creates, as discussed in this blog post of hers
Thus overlapping segments were not part of the initial design. However I can see people want to use it to look at other CSV files, so I am adding some features to accommodate and considering how to show overlaps other than by assigning them to different sides.
Here is what the output looks like now (click for a larger version):
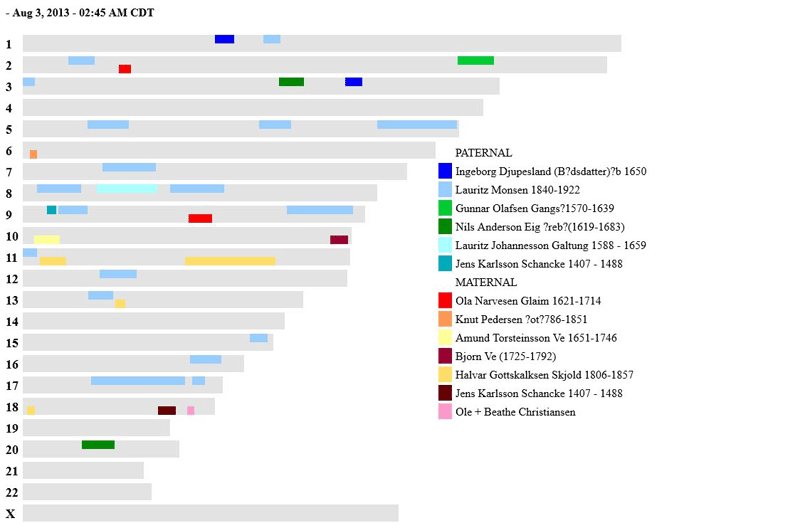
Hi, thank you for doing this. 🙂
Thanks for putting this together! I can’t wait to get a few minutes to try it out. Would it perhaps be more accurate to label the MRCA column as MRCAC (most recent common ancestral couple) since there is always a couple and not just one individual that we trace back to when using autosomal DNA?
You can label the MRCA column anything you want, just tell the program what it is called
And I have numerous cases where I know which person the DNA came from in a couple because the relative I share DNA with was descended from a different marriage!
Hi, this seems to be great. Suggestion: could you still add there a third class, both. In many cases there are DNA-relatives that are one’s relatives from both Mom’s and Dad’s side at the same time. Letter B & one color, something like that.
both is on the feature list for either the next release or the one after
There was an occasional bug with segments that started below about 100,000 which is now fixed. Thank you Trevor!
Thanks very much for the fix. All I need now, to make better use of the Tool, is to improve on my zero confirmed matches at 23andMe and Ancestry DNA, and my three confirmed matches at FTDNA Family Finder.
LOL Trevor. I have been doing this for a year now and have found about ten new confirmed Norwegian 4th-7th cousins but zero german (jewish) confirmed.
But what I have been doing is getting as many known 2nd-5th cousins to test as I can to narrow down where specific segments come from. Also postulated relatives. I have talked two 3rd cousins, two second cousins, one 4th and many 5th into doing the test! I find seeing how DNA inheritance works in my own family fascinating. Of course having two completely different ethnic sides makes it easier as well.
And yes I have tested at both 23andme and family tree DNA.
I have problem, this is my test data:
Side,Chr,Start point,End point,cMs,MRCA
P,2,86.0,100.6,5.9,Alex Whyte
P,4,32.5,38.0,5.9,Alisa Tongg
P,11,79.0,87.4,7.3,ALLI B
but I got the message
Column heading side is required.
–> Column heading MRCA is required.
Please fix your CSV and try again
Kind regards
Åke Sjödin
Sweden
The column headings muct be the first line of the file, that being said …
Mac users are often having this problem because the program they use does not create the line endings in a way that is compatible with a PC. One of my Mac users said
“I switched to using Numbers (Mac) rather than Open Office which then gave me the option to save as csv fie and worked like a charm.”
On a PC you could read the CSV file in notepad, add a dummy column e.g. ,dummy and then save it to fix line endings
Mac users are having problems because Microsoft Excel for Mac 2011 saves CSV files using only carriage returns (CR) for line breaks, which hasn’t been used since MacOS 9. MacOS X has used the standard Unix convention of line feeds (LF) for line breaks since 2001. In Excel 2011, there are three options for CSV files in the Save As dialog (among other formats): Comma Separated Values (.csv), Windows Comma Separated (.csv), and MS-DOS Separated Values (.csv). I just saved a file as all three, and only the Windows format saves the file with CRLF for line breaks, which is strange because that’s also what DOS always used.
I also tested Numbers ’09, which likewise has three options when exporting to CSV, but all three use CRLF. So Mac users can either use Numbers, make sure to save as “Windows Comma Separated (.csv)” in Excel if they don’t have Numbers, or they could open the CSV file in a text editor like TextWrangler, which can easily change the line breaks to “Windows (CRLF)”.
You could also make it cross platform on the server. See the note under “Return Values” in the php documentation for fgetcsv(). In the php.ini configuration file, auto_detect_line_endings is off by default. Turning it on should make the problem irrelevant.
Thanks for the help
/Åke
Thanks for this 😉
Limiting to 10 ancestors on each side is a bit…limiting.
Great mapping tool, though.
Thanks
/Åke
So should I repeat the colors? Add some grays? It is difficult to make the colors distinct and usable even for people with visual disabilities. Hard to add more of them. What I am considering is allowing you to specify the colors, so unlimited in that case
The original idea was to show what DNA came from which ancestor so 8 to a side seemed plenty. Thus gg-grandparent level, but it seems there is a desire to use the tool for looking at lots of matches
I suspected that was the hangup.
Your color scheme is great for smaller numbers of relatives. Limiting my map to great grandparents generated very nice results. I like the blue/green/etc. for paternal matches and red/yellow/etc. for maternal ones. That makes it very easy to process and conceptualize visually.
I suppose you could add black and white.
For large numbers of matches I suppose you might also separate sister chromosomes so that all colors could be used for both sides. Not sure that would be a satisfactory solution, but if I have any other bright ideas, I’ll be sure to chime in.
Oh, and I do like the idea for user-specified colors.
Looks like a cool tool i have a windows computer but have no clue how to use tool 🙁
You need a spreadsheet of matches that you want to plot. Read this article
http://blog.kittycooper.com/2012/11/making-a-spreadsheet-of-your-dna-matches/
I haven’t worked with spreadsheets for 20 years. Your instructions say to put side,Start point,End point,cMs, MRCA in line 1.
I did exactly that and I get this message.
Column heading Chromosome is required.
–> Column heading Start is required.
–> Column heading End is required.
–> Column heading side is required.
–> Column heading MRCA is required.
–> Column heading centiMorgans (cM) is required.
Please fix your CSV and try again
What do I do?
I found it, Excel had saved my file as an XLS file.
I am having a problem. I changed my file to a CSV file and made sure my headings fit with what was requied but all I get as a result is a blank chromosome chart. What am I doing wrong?
Michelle
It only maps the lines which have something in both the side column and the MRCA column.
Thank you!! A wonderful tool and I’m glad I can stop doing this by hand now!
I noticed for my segments that have a start position <1,000,000 they don't display. For example one of my segments starts at 213,000 and ends at 35,000,000. It doesn't display. But if I change the start point to "1" it will display.
Perhaps this is a bug?
It seemed like if the start point is over 1 million, it works fantastically however.
Gee I thought that early bug was fixed, but it looks like I only fixed it for less than 100,000. I will email you for a copy of your CSV to test with if you would be so kind
OK we figured this out, not the bug it once had … Erik’s start and stop numbers were saved in text format, so had quotes around them and commas in them. I will try to add the feature to the tool to handle this case.
In the meanwhile everyone be careful how you format the numbers columns in your spreadsheet!! When I just cut and paste or use downloaded CSVs the columns have been fine for me. You can click at the top of the column and then on format cells to be sure they are formatted as numbers in OpenOffice
I converted an Excel 2007 file to a MS-DOS CVS file and made sure the headings matched exactly with the requirements shown (Caps as indicated) and it worked perfectly.
I have updated this post with the enhancements I hope to add in the next few weeks and a link to the documentation which is permanently in the top menu. I think it is greatly improved and I am hoping for less problems now due to user error. Contributions to the documentation are also very welcome.
I am confused. I uploaded my csv and it says i don’t have a csv file. I don’t think the colunns are right but I downloaded it from 23andme. I don’t want to alter anything because I might screw it up, Excel is not my friend….
Lynn you cannot use the CSV downloaded from 23andme without making changes. Make a copy of the file. Look at it in Excel and add the needed columns lsited above, side and MRCA.
Mind you this tool was not written to display all the matches from 23andme. It was written to display the people’s segments that you specify, where you know how you are related. Another version of the tool will be out soon which will make a more generalized mapping easier to do.
Many of you seemed to want a tool to map overlapping DNA segments from unknown relatives rather than the map of known segments. So I made a new tool for that purpose and called it a DNA segment mapper http://blog.kittycooper.com/tools/segment-mapper/
My #2, #3, and #4 matches from 23andme are not showing up here at all. Can you tell me why that might be? Thanks!
Kath
This tool is for known ancestral bits of DNA and only shows up to 20 ancestors on each side (you can have more if you specify your own colors). So if these matches are listed towards the end of your file they might not show up.
You may want to be using the segment mapper and I recommend sorting by segment size, largest to smallest, to get your best matches to show. See http://blog.kittycooper.com/tools/segment-mapper/
Thanks for explaining.
I want to check something with you. People who overlap on the segment mapper are not necessarily related to each other — correct? That is, they could be on opposite sides of the family? (They are not sorted into families? I don’t see anything in the data that would provide that information, but I might have missed something.)
And thanks again! I will never do it “the old way” again! (Well, maybe when I look at ethnicity — sigh.)
Kath, you are correct. Overlapping does not mean they match.
You need to compare them to each other to see if they match which you can do on GEDmatch or 23andme in FIA. At family tree DNA you can see if they are ICW but you need to ask one of them if they match the other.
See if this post helps http://blog.kittycooper.com/2014/10/when-is-a-dna-segment-match-a-real-match-ibd-or-ibs-or-ibc/
How do you trace DNA back to a single MRCA when, unless descended from the products of different marriages, we will have TWO MRCAs (e.g. gg grandparents) for each match?
Ali, you are correct, usually it is an ancestral couple who you share relatives with, so for mapping I usually put in the name of the child of that couple whom I descend from. Of course there are also cases of multiple husbands or wives where I actually know which person in the couple gave me that piece of DNA.
My tools are meant for people to use as they wish so you could put the couple in instead.