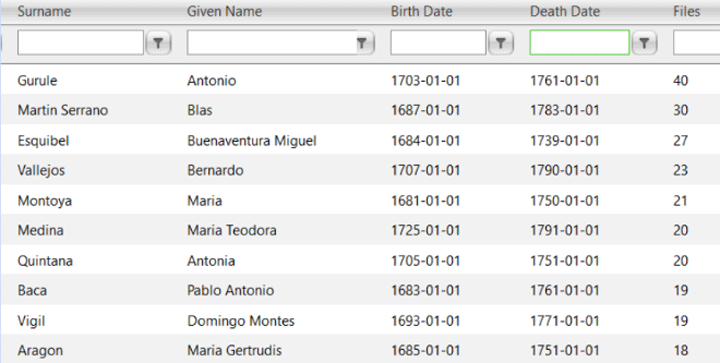
It is difficult to solve adoption cases in endogamous communities because everyone will share the same 4th and 5th grandparents, often multiple times, so the methodology for finding a birth parent from 3rd and 4th cousin matches just does not work. You have to wait for a few second cousin or closer matches.
Tessa was looking for her unknown biological father. Her mother had given her a name, Rudy Padilla, and said he was perhaps Mexican. I ran a GWorks for her which I showed in my lecture about unknown parentage at the SCGS Jamboree. This is the full story.
I had never seen ancestors who were in 30-40 trees before! How can that be? Perhaps endogamy? Then I looked at the names and recognized many of the surnames. These are the Spanish soldiers who were among the earliest European settlers of New Mexico.
These soldiers who came to the Southwest in the 1600s and 1700s mostly had to take Pueblo women as brides or not get married. A few brought wives with them from Mexico of presumed Spanish descent. For many years these Spanish “first families” of New Mexico hid the native part of their roots. Now many are proud of this heritage. Click here for an article about that which mentions the New Mexican woman in those Ancestry ads who discovered her Native American roots with DNA. By the way, Tessa shows 17% Native American at Ancestry
.
I told Tessa that success finding her dad could take a very long time since she would need to wait for close matches, but to please upload to MyHeritage and Family Tree DNA to look for more relatives. She had tested at both 23andMe and Ancestry DNA.
Continue reading