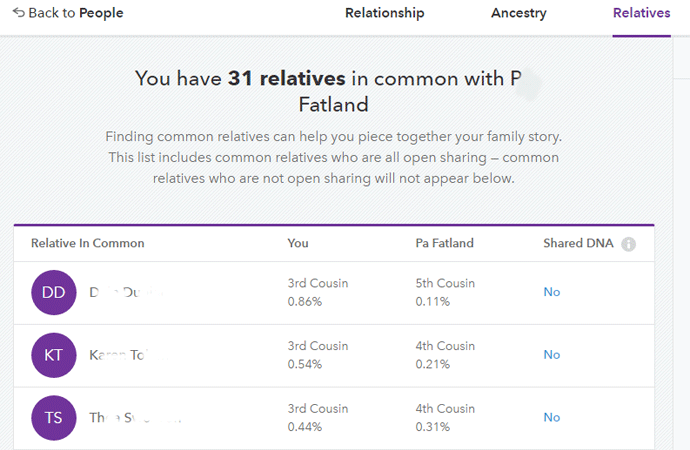
An exciting triangulation feature is available at 23andme. This feature shows you the relatives you have in common with one other profile and whether or not they triangulate with the two of you.
To use this tool you go to Family & Friends > DNA relatives and pick a DNA relative to look at. Scroll down the page towards the bottom where you will see a display like this (names removed for privacy): [previous 2 paragraphs updated 10 May 2019]
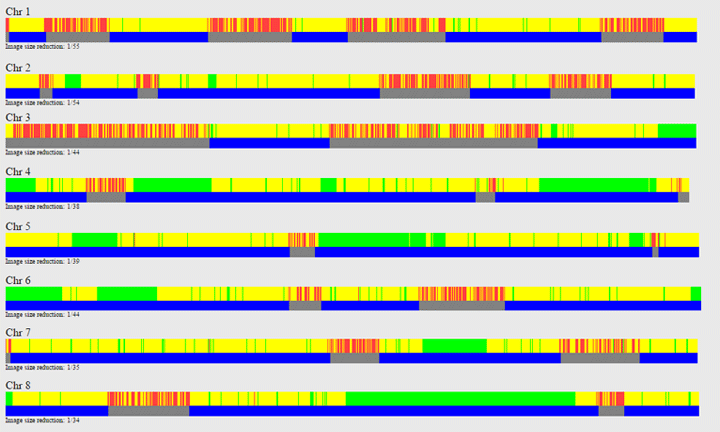
When there is a “Yes” in the right hand column, there is a DNA segment in common between the two people you are comparing and this third person thus you triangulate. A “No” means that although you each share DNA with this relative, there is no segment of DNA where all three of you match. Click on the Yes (or a No) to see the three of you compared in the chromosome browser.
Once in the chromosome browser, you can use the “Update View – Edit” to add more people to the comparison with the first person. This is the same chromosome browser that you get to from Tools > DNA relatives > DNA
Jason Lee has done a nice write up with more details on his DNA blog at Tumblr:
http://dnagenealogy.tumblr.com/post/144966221253/relatives-in-common-quick-reaction