There have been so many claims in the alternate medicine arena about health problems caused by variations in the MTHFR gene that I was not surprised to get this request from a favorite cousin:
Could you please check to see if there is information in my Ancestry
test that tells you whether I have a bad version of the MTHFR gene?
Like many genes, the MTHFR is made up of a long DNA chain including many SNPs (Single-nucleotide polymorphisms), pronounced “snip.” SNPs are places where a single letter in the DNA code often changes to another letter. Since these can vary from one person to another they are useful for figuring out ethnicity. However some variations can have health effects. Typically you would need more than one variation to greatly increase your risks of specific diseases, but not always.
So what does the MTHFR gene do? It has the instructions for making an enzyme critical to turning the amino acid homocysteine to another amino acid, methionine, a building block for making proteins. That is a simplification; click here for the full explanation from the National Library of Medicine (NIH).
One health condition, known as homocystinuria, causing blood homocysteine levels to be too high, is caused by variations in this gene. However that can easily be addressed with certain vitamin B supplements. Geneticist Charis Eng discusses why a DNA test is not needed to diagnose or treat this at https://health.clevelandclinic.org/a-genetic-test-you-dont-need/
The genetic cause is not simple, according to the NIH at https://ghr.nlm.nih.gov/gene/MTHFR#conditions – “At least 40 mutations in the MTHFR gene have been identified in people with homocystinuria, a disorder in which the body is unable to process homocysteine and methionine properly.”
So can I answer my cousin’s question? There are several SNPs in the MTHR that have been intensely studied, maybe these were tested in her Ancestry.com test.
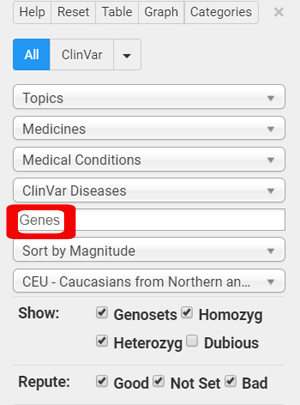
My advice to her was to upload to Promethease.com which will analyze this nicely for her. When you look at the report, type MTHFR in the box labeled Genes (outlined in red in my image here) and let it tell you your risks.
Of course I still had to figure out whether I could find the most interesting MTHFR SNPs in the raw results. If they are not there, then Promethease will not be much use.
Another place that discusses the MTHFR variants is the NIH rare diseases web site which says “These variants are common. In America, about 25% of people who are Hispanic, and 10-15% of people who are Caucasian have two copies of C677T.”
and “Studies have found that women with two C677T gene variants have an increased risk for having a child with a neural tube defect. Studies have also found that men and women with two C677T gene variants and elevated homocysteine levels may be at a mild increased risk for blood clots (venous thromboembolism).”
C677T is the one to look for then, but it was not immediately obvious how to turn that into a location listed in her Ancestry DNA results. That notation means that a C has changed to a T at location 677 in the gene, but it does not place it in the genome. Having two of these may cause various problems.
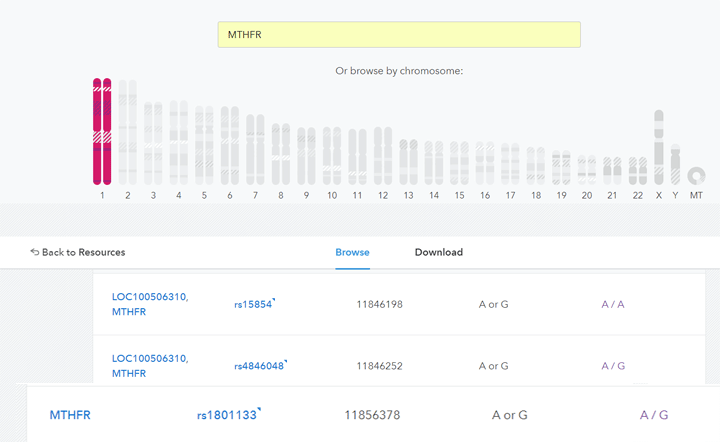
SNPedia.com is my go to website for explaining DNA topics in close to plain English. Sure enough, when I searched for MTHFR there I found a page which translated those numbers to the rsid numbers in the raw DNA data file. According to https://www.snpedia.com/index.php/MTHFR
There are 3 common SNPs giving rise to MTHFR alleles:
rs1801133, also known as C677T or A222V
rs1801131, also known as A1298C or E429A
rs2274976, also known as G1793A or R594Q
also I learned from the page about that SNP – https://www.snpedia.com/index.php/Rs1801133 that “Homozygous rs1801133(T;T) individuals have ~30% of the expected MTHFR enzyme activity, and rs1801133(C;T) heterozygotes have ~65% activity, compared to the most common genotype, rs1801133(C;C).”
Once I had the rsid number, searching the text file of the raw data for that number was a simple matter and both rs1801113 and rs1801133 were there. Of course it is showing a G not a C (they are paired so they are basically equivalent in the results just like A and T are).
Out of curiosity, I used the browse raw data (under tools) at 23andme to check if my 2011 test had those SNPs and if a cousin on the v4 chip from 2015 had them. Yes both tests had these SNPs but I hear that the latest tests may not have them.
23andme wrote a blog post about the MTHFR gene a little over a year ago and they say: “Some websites have spread the idea that having one or two copies of an MTHFR variant can lead to dozens of negative health consequences. There are a couple problems with this claim. First, it’s unlikely that variants in a single gene could cause dozens of unrelated health problems. Second, the C677T and A1289C variants are very common: in some ethnicities, more than 50 percent of people have at least one copy of one of these variants. Most disease-causing genetic variants are not this common.
SNPedia has this common sense summary about MTHFR :
The “only scientific consensus about the impact of MTHFR variants is that certain very rare mutations can cause a disease called Homocystinuria. How rare are these mutations? Less than 1 person in a million has homocystinuria due to MTHFR mutations. We know you are unique, but you are very, very unlikely to be that 1 person in a million.
The questions we most commonly receive are not about these rare mutations; instead, they are about the very common variants (aka polymorphisms). The variants are usually called C677T and A1298C; in SNPedia, these are represented by rs1801133 and rs1801131, respectively.
There is scientific consensus about these common variants as well. The consensus is that (1) there is no medical reason to test these common variants, and (2) there no recommended actions based on the results of such genetic tests.”
In conclusion, genetics is far more complicated than a single variation in a single gene causing a problem, but disclaimer, I am not a geneticist, just an interested citizen scientist.
I hope this answers your question my beloved cousin.
Great post!
Also worth keeping in mind for many traits that are not caused by a single mutation (i.e. most traits and diseases) – the presence of a single risk allele (the “bad” version of a SNP) can mean different things in different people. Reported effects of risk alleles are largely based on averages across many people.
The effects of some SNP variants can vary based on environmental factors, genotypes at other genes, sex, or even ethnic backgrounds – and many of these interactions may not yet be known or understood.
Nature’s Scitable website has some overviews of these more complicated concepts, including “Epistasis: Gene Interaction and Phenotype Effects”, “Environmental Influences on Gene Expression”, and “Phenotype Variability: Penetrance and Expressivity.”
Thanks for the very informative comment! What an interesting site Nature has put together, I like this article about epistasis (where genes can mask and alter the effects of other genes)
https://www.nature.com/scitable/topicpage/epistasis-gene-interaction-and-the-phenotypic-expression-907