Alert, there is a bug in my tool to convert text files to GEDCOMs: very large Ahnentafel numbers like “46406041600” will cause it to hang.
I will add code to ignore large numbers by May (end of this week). If you are a regular user of this tool, check this post for the update when the new feature is released.
Something must have changed on the collection of trees because I had three emails in the last week complaining that my tool hung and did not complete the conversion. In all cases, the Ahnentafel went up to extremely large numbers, so eliminating those last few lines fixed the problem
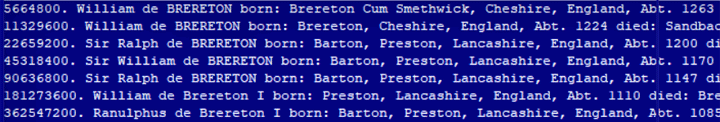
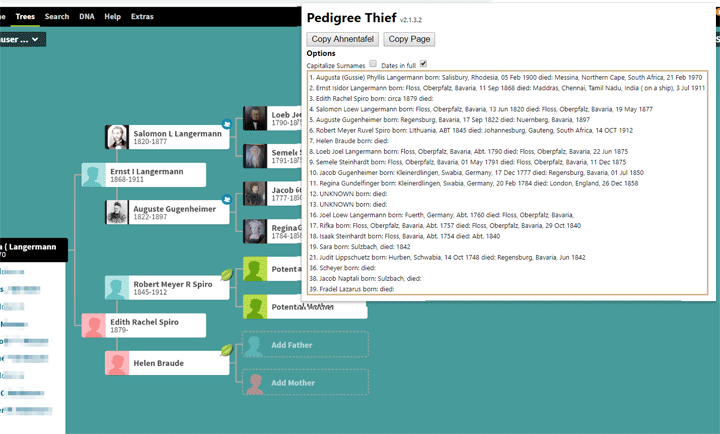
Here is an example of the last several lines of a file that did not work:
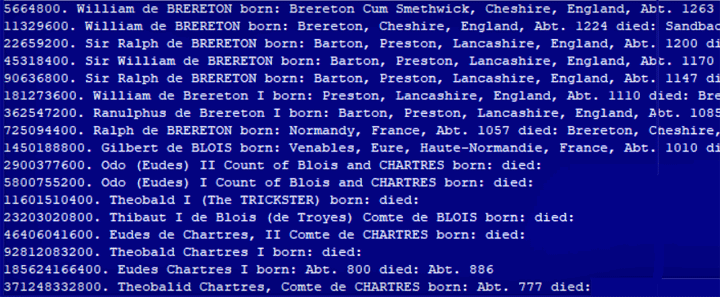
Here are the last few lines of the same file after removing the lines with very high numbers. This version worked.
Do you really need these people born in the 1200s? What is the probability that they even are actually your ancestors?
This appears to be some sort of limitation in either the storage space for the program or the number sizes. Thus I propose to modify the code to ignore ahnentafel numbers with more than seven digits and to have it tell you that it did that.
Any other ideas out there? Remember I make almost no money on this, just the occasional small thank you donation, so I am not looking for a solution that will take lots of my time.